6J38
 
 | | Crystal structure of CmiS2 | | Descriptor: | FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2019-01-04 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6K97
 
 | | Crystal structure of fusion DH domain | | Descriptor: | Fusion DH, SULFATE ION | | Authors: | Kawasaki, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional and Structural Analyses of the Split-Dehydratase Domain in the Biosynthesis of Macrolactam Polyketide Cremimycin.
Biochemistry, 58, 2019
|
|
6K96
 
 | | Crystal structure of Ari2 | | Descriptor: | Five-membered-cyclitol-phosphate synthase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Miyanaga, A, Tsunoda, T, Kudo, F, Eguchi, T. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereochemistry in the Reaction of themyo-Inositol Phosphate Synthase Ortholog Ari2 during Aristeromycin Biosynthesis.
Biochemistry, 58, 2019
|
|
6J39
 
 | | Crystal structure of CmiS2 with inhibitor | | Descriptor: | (3R)-3-[(carboxymethyl)sulfanyl]nonanoic acid, FAD-dependent glycine oxydase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawasaki, D, Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2019-01-04 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Analysis of the Glycine Oxidase Homologue CmiS2 Reveals a Unique Substrate Recognition Mechanism for Formation of a beta-Amino Acid Starter Unit in Cremimycin Biosynthesis.
Biochemistry, 58, 2019
|
|
5ZK4
 
 | | The structure of DSZS acyltransferase with carrier protein | | Descriptor: | DisA protein, DisD protein, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Miyanaga, A, Ouchi, R, Kudo, F, Eguchi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of protein-protein interactions between a trans-acting acyltransferase and acyl carrier protein in polyketide disorazole biosynthesis
J. Am. Chem. Soc., 140, 2018
|
|
6AKD
 
 | | Crystal structure of IdnL7 | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, AMP-dependent synthetase and ligase, GLYCEROL | | Authors: | Cieslak, J, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-08-31 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of IdnL7, an adenylation enzyme involved in incednine biosynthesis.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WMR
 
 | | Crystal structure of VinJ | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, Proline iminopeptidase | | Authors: | Shinohara, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the amidohydrolase VinJ shows a unique hydrophobic tunnel for its interaction with polyketide substrates
Febs Lett., 588, 2014
|
|
3WV4
 
 | | Crystal structure of VinN | | Descriptor: | Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
3WVN
 
 | | Complex structure of VinN with L-aspartate | | Descriptor: | ASPARTIC ACID, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-30 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
6JW7
 
 | | The crystal structure of KanD2 in complex with NADH and 3"-deamino-3"-hydroxykanamycin A | | Descriptor: | (2R,3S,4S,5R,6R)-2-(aminomethyl)-6-[(1R,2S,3S,4R,6S)-4,6-bis(azanyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-oxidanyl-cyclohexyl]oxy-oxane-3,4,5-triol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
7DQ5
 
 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
6JW8
 
 | | The crystal structure of KanD2 in complex with NADH and 3"-deamino-3"-hydroxykanamycin B | | Descriptor: | (2S,3R,4S,5S,6R)-2-[(1S,2S,3R,4S,6R)-3-[(2R,3R,4R,5S,6R)-6-(aminomethyl)-3-azanyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-4,6-bis(azanyl)-2-oxidanyl-cyclohexyl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
6JW6
 
 | | The crystal structure of KanD2 in complex with NAD | | Descriptor: | Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
7EIQ
 
 | |
7EIR
 
 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 6S | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, GLYCEROL, ... | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
7EIP
 
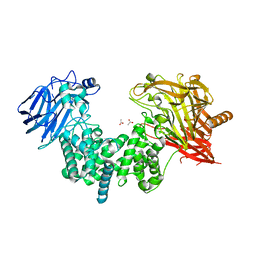 | | Crystal structure of ligand-free chondroitin ABC lyase I | | Descriptor: | ACETATE ION, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
7EIS
 
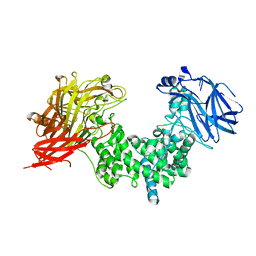 | |
7F2R
 
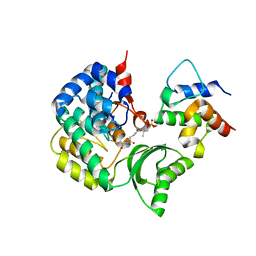 | | Crystal structure of VinK-VinL covalent complex formed with a pantetheineamide cross-linking probe | | Descriptor: | Acyl-carrier-protein, Malonyl-CoA-[acyl-carrier-protein] transacylase, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Miyanaga, A, Ouchi, R, Kudo, F, Eguchi, T. | | Deposit date: | 2021-06-14 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex structure of the acyltransferase VinK and the carrier protein VinL with a pantetheine cross-linking probe.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7CL2
 
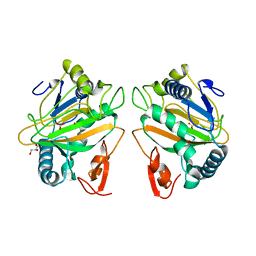 | | The crystal structure of KanJ | | Descriptor: | GLYCEROL, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
7CL5
 
 | | The crystal structure of KanJ in complex with kanamycin B and N-oxalylglycine | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Kanamycin B dioxygenase, N-OXALYLGLYCINE, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
7CL4
 
 | | The crystal structure of KanJ in complex with N-oxalylglycine | | Descriptor: | Kanamycin B dioxygenase, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
7CL6
 
 | | The crystal structure of KanJ in complex with neamine and N-oxalylglycine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, Kanamycin B dioxygenase, N-OXALYLGLYCINE, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
7CL3
 
 | | The crystal structure of KanJ in complex with kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
