6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | 分子名称: | Peptidoglycan endopeptidase RipA | | 著者 | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | 登録日 | 2017-11-07 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
2XFY
 
 | | Crystal structure of Barley Beta-Amylase complexed with alpha- cyclodextrin | | 分子名称: | 1,2-ETHANEDIOL, BETA-AMYLASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose) | | 著者 | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | 登録日 | 2010-05-28 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.207 Å) | | 主引用文献 | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
6EZN
 
 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | 分子名称: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-17 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
2XGI
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3,4- epoxybutyl alpha-D-glucopyranoside | | 分子名称: | (3R)-3-hydroxybutyl alpha-D-glucopyranoside, (3S)-3-hydroxybutyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, ... | | 著者 | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | 登録日 | 2010-06-04 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley beta-amylase.
Mol Biosyst, 7, 2011
|
|
4Y74
 
 | | Yeast 20S proteasome in complex with Ac-LAL-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAL-ep, CHLORIDE ION, ... | | 著者 | Groll, M, Huber, E.M. | | 登録日 | 2015-02-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
2XG9
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D- glucopyranosylmoranoline | | 分子名称: | 1,2-ETHANEDIOL, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | 著者 | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | 登録日 | 2010-06-02 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
4Y8M
 
 | | Yeast 20S proteasome beta7-delta7_Cter mutant | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
2XLQ
 
 | | Structural and Mechanistic Analysis of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway | | 分子名称: | (2R)-2-HYDROXY-3-(4-HYDROXYPHENYL)PROPANOIC ACID, CLOQ, FORMIC ACID | | 著者 | Metzger, U, Keller, S, Stevenson, C.E.M, Heide, L, Lawson, D.M. | | 登録日 | 2010-07-21 | | 公開日 | 2010-10-27 | | 最終更新日 | 2019-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase Cloq from the Clorobiocin Biosynthetic Pathway.
J.Mol.Biol., 404, 2010
|
|
6F9Q
 
 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | 分子名称: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | 登録日 | 2017-12-15 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | 分子名称: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | 著者 | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | 登録日 | 2010-06-02 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
4Y70
 
 | | Yeast 20S proteasome in complex with Ac-LAV-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAV-ep, CHLORIDE ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y8P
 
 | |
4Y9Y
 
 | | Yeast 20S proteasome beta2-H116E mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-17 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | 分子名称: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | 著者 | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | 登録日 | 2017-12-13 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
4YLS
 
 | | Tubulin Glutamylase | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tubulin polyglutamylase TTLL7 | | 著者 | Garnham, C.P, Vemu, A, Wilson-Kubalek, E.M, Yu, I, Szyk, A, Lander, G.C, Milligan, R.A, Roll-Mecak, A. | | 登録日 | 2015-03-05 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases.
Cell, 161, 2015
|
|
6F96
 
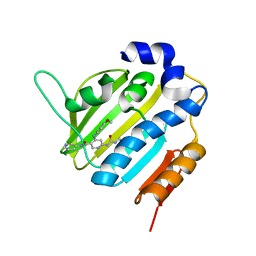 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | 分子名称: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | 著者 | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
8EHK
 
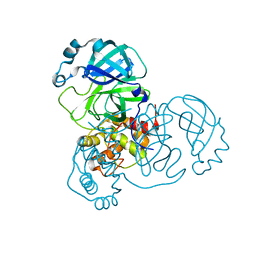 | |
8EHM
 
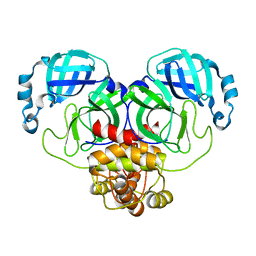 | |
8EHL
 
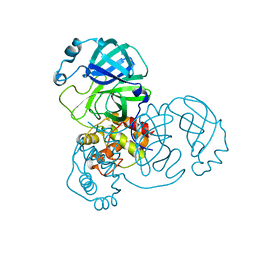 | |
8EHJ
 
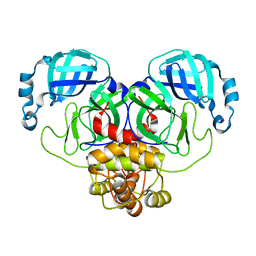 | |
4UZ2
 
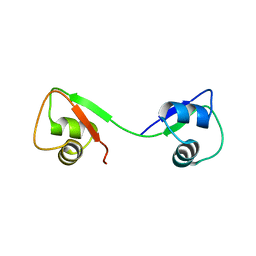 | |
2Y2Z
 
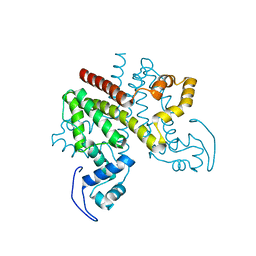 | | ligand-free form of TetR-like repressor SimR | | 分子名称: | PUTATIVE REPRESSOR SIMREG2 | | 著者 | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | 登録日 | 2010-12-17 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
4YLR
 
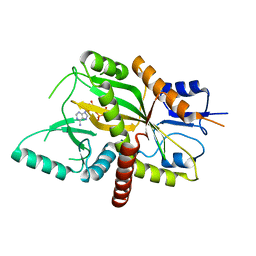 | | Tubulin Glutamylase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Tubulin polyglutamylase TTLL7 | | 著者 | Garnham, C.P, Vemu, A, Wilson-Kubalek, E.M, Yu, I, Szyk, A, Lander, G.C, Milligan, R.A, Roll-Mecak, A. | | 登録日 | 2015-03-05 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases.
Cell, 161, 2015
|
|
4WUB
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | 分子名称: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | 著者 | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | 登録日 | 2014-10-31 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | 分子名称: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | 著者 | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | 登録日 | 2017-12-12 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
