6XAV
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
2Y5K
 
 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | 分子名称: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | 著者 | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | 登録日 | 2011-01-14 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7NG3
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
3UFX
 
 | | Thermus aquaticus succinyl-CoA synthetase in complex with GDP-Mn2+ | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, Succinyl-CoA synthetase beta subunit, ... | | 著者 | Fraser, M.E. | | 登録日 | 2011-11-01 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Biochemical and structural characterization of the GTP-preferring succinyl-CoA synthetase from Thermus aquaticus.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7NG6
 
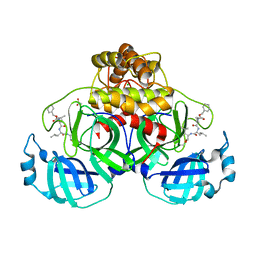 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | 分子名称: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
1VHR
 
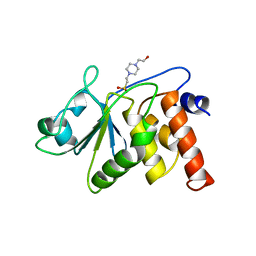 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE VHR, SULFATE ION | | 著者 | Yuvaniyama, J, Denu, J.M, Dixon, J.E, Saper, M.A. | | 登録日 | 1996-02-20 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the dual specificity protein phosphatase VHR.
Science, 272, 1996
|
|
5KWQ
 
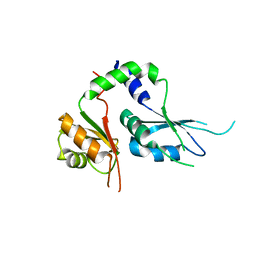 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | 分子名称: | Poly(U)-binding-splicing factor PUF60 | | 著者 | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | 登録日 | 2016-07-18 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7NF5
 
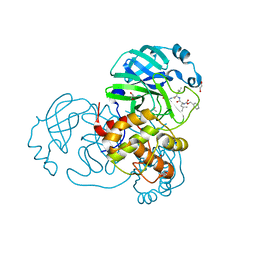 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-05 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
5W4K
 
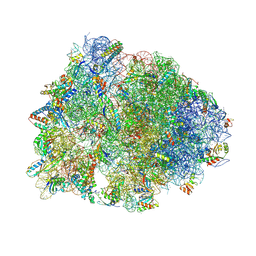 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | 登録日 | 2017-06-12 | | 公開日 | 2017-08-30 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
2GHD
 
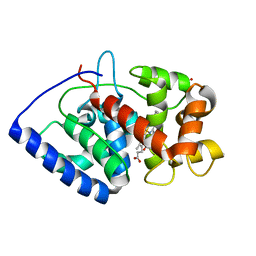 | | Conformational mobility in the active site of a heme peroxidase | | 分子名称: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | 著者 | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | 登録日 | 2006-03-27 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
1W6V
 
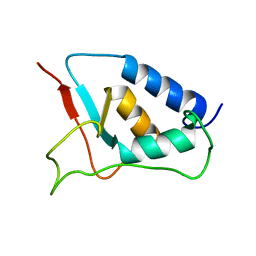 | | Solution structure of the DUSP domain of hUSP15 | | 分子名称: | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | 著者 | De Jong, R.D, Ab, E, Diercks, T, Truffault, V, Daniels, M, Kaptein, R, Folkers, G.E. | | 登録日 | 2004-08-24 | | 公開日 | 2006-01-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Human Ubiquitin-Specific Protease 15 Dusp Domain.
J.Biol.Chem., 281, 2006
|
|
4OND
 
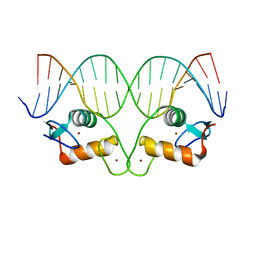 | | Ancestral Steroid Receptor 2 DBD helix mutant - ERE DNA complex | | 分子名称: | 5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*GP*AP*GP*TP*GP*AP*CP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*GP*TP*CP*AP*CP*TP*CP*TP*GP*AP*CP*CP*TP*G)-3', Ancestral SR2 Helix Mutant, ... | | 著者 | Ortlund, E.O, Murphy, M.N. | | 登録日 | 2014-01-28 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.253 Å) | | 主引用文献 | Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module.
Cell(Cambridge,Mass.), 159, 2014
|
|
3UKP
 
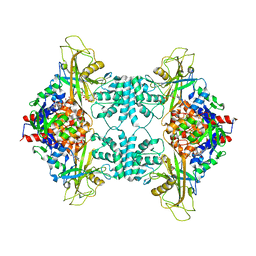 | |
2GGN
 
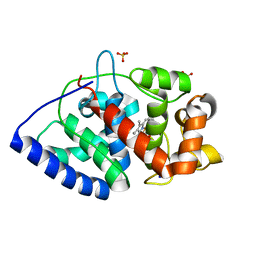 | | Conformational mobility in the active site of a heme peroxidase | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, SULFATE ION, ... | | 著者 | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | 登録日 | 2006-03-24 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
2Y54
 
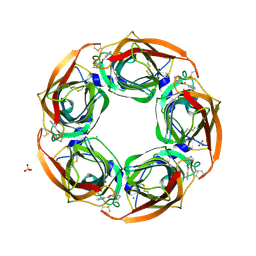 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Fragment 1) | | 分子名称: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | 著者 | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | 登録日 | 2011-01-12 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
3KVV
 
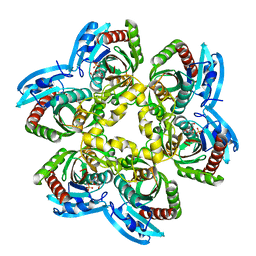 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | 分子名称: | 1,4-anhydro-D-erythro-pent-1-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | 著者 | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | 登録日 | 2009-11-30 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
6XP0
 
 | |
3UC1
 
 | |
3UKH
 
 | |
3UNC
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | 分子名称: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | 著者 | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | 登録日 | 2011-11-15 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
2XJZ
 
 | | Crystal structure of the LMO2:LDB1-LID complex, C2 crystal form | | 分子名称: | CHLORIDE ION, LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ... | | 著者 | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | 登録日 | 2010-07-06 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
6XAS
 
 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex | | 分子名称: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | 登録日 | 2020-06-04 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
6XJA
 
 | |
2GHK
 
 | | Conformational mobility in the active site of a heme peroxidase | | 分子名称: | CYANIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | 登録日 | 2006-03-27 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
5DN6
 
 | | ATP synthase from Paracoccus denitrificans | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F0 subcomplex C subunit, ... | | 著者 | Morales-Rios, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | 登録日 | 2015-09-09 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.98 Å) | | 主引用文献 | Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
