1FSE
 
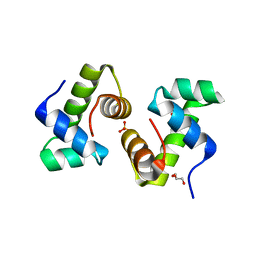 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS REGULATORY PROTEIN GERE | | Descriptor: | GERE, GLYCEROL, SULFATE ION | | Authors: | Ducros, V.M.-A, Lewis, R.J, Verma, C.S, Dodson, E.J, Leonard, G, Turkenburg, J.P, Murshudov, G.N, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2000-09-08 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis.
J.Mol.Biol., 306, 2001
|
|
1OJJ
 
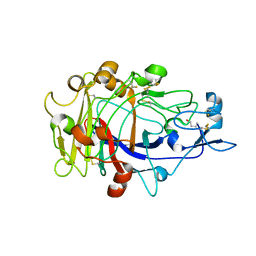 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJK
 
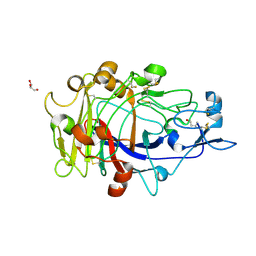 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJI
 
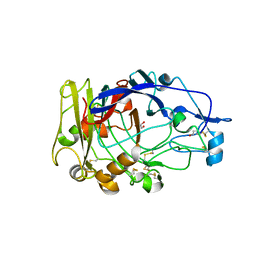 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1UWR
 
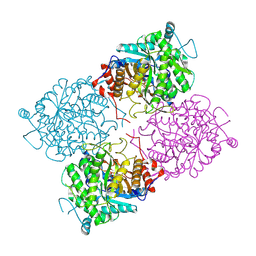 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
1UWT
 
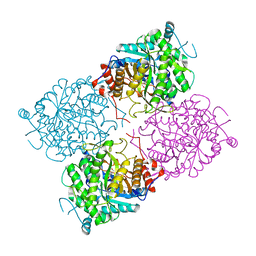 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-galactohydroximo-1,5-lactam | | Descriptor: | (2E,3R,4R,5R,6S)-3,4,5-TRIHYDROXY-6-(HYDROXYMETHYL)-2-PIPERIDINONE, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
1UWS
 
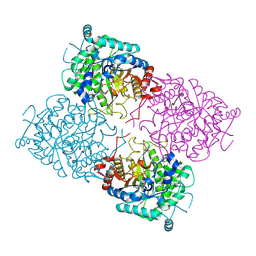 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-glucose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
1UWQ
 
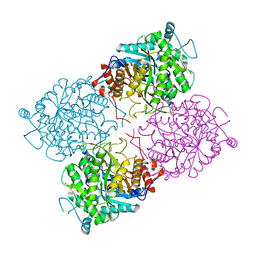 | | Structure of beta-glycosidase from Sulfolobus solfataricus | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
1UWU
 
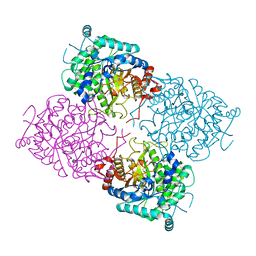 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-glucohydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
2VX5
 
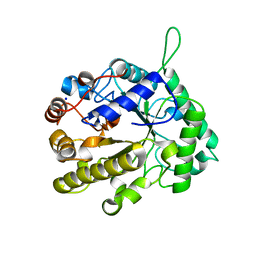 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C MANNOSE-BOUND FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, SODIUM ION, beta-D-mannopyranose | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
2VX6
 
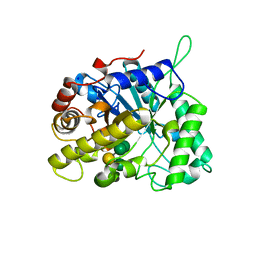 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C Gal1Man4-BOUND FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, SODIUM ION, beta-D-mannopyranose-(1-4)-[alpha-D-galactopyranose-(1-6)]beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
2VX7
 
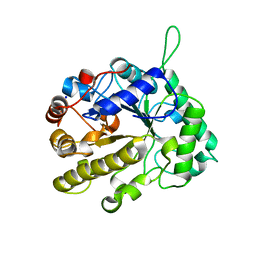 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C MANNOBIOSE-BOUND FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, SODIUM ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
2VX4
 
 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C NATIVE FORM | | Descriptor: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, GLYCEROL, SODIUM ION | | Authors: | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
