3HL3
 
 | | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose. | | Descriptor: | Glucose-1-phosphate thymidylyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose.
TO BE PUBLISHED
|
|
3INP
 
 | | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, D-ribulose-phosphate 3-epimerase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Scott, P, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis.
TO BE PUBLISHED
|
|
3KY7
 
 | | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252 | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, See, R, Zoraghi, R, Reiner, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252
To be Published
|
|
3ROH
 
 | | Crystal Structure of Leukotoxin (LukE) from Staphylococcus aureus subsp. aureus COL. | | Descriptor: | CHLORIDE ION, Leucotoxin LukEv, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3LXM
 
 | | 2.00 Angstrom resolution crystal structure of a catalytic subunit of an aspartate carbamoyltransferase (pyrB) from Yersinia pestis CO92 | | Descriptor: | Aspartate carbamoyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of a catalytic subunit of an aspartate carbamoyltransferase (pyrB) from Yersinia pestis CO92
To be Published
|
|
3SD7
 
 | | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative phosphatase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile.
TO BE PUBLISHED
|
|
4O0N
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5SV5
 
 | | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis. | | Descriptor: | Microbial collagenase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom Crystal Structure of pre-Peptidase C-terminal Domain of Collagenase from Bacillus anthracis.
To Be Published
|
|
5TKW
 
 | | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae. | | Descriptor: | FORMIC ACID, Type II secretion system protein L | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.
To Be Published
|
|
5TY0
 
 | | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila. | | Descriptor: | Elongation factor G, SODIUM ION, beta-D-glucopyranose | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Crystal Structure of N-terminal Fragment (residues 1-419) of Elongation Factor G from Legionella pneumophila.
To Be Published
|
|
5TSE
 
 | | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii. | | Descriptor: | FORMIC ACID, LPS-assembly lipoprotein LptE | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.
To Be Published
|
|
5U4Q
 
 | | 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-glucose 4,6-dehydratase | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-05 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Resolution Crystal Structure of NAD-Dependent Epimerase from Klebsiella pneumoniae in Complex with NAD.
To Be Published
|
|
5TRO
 
 | | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.
To Be Published
|
|
5TU0
 
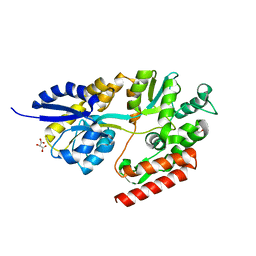 | | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose | | Descriptor: | Lmo2125 protein, TARTRONATE, TRIETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose.
To Be Published
|
|
5TV2
 
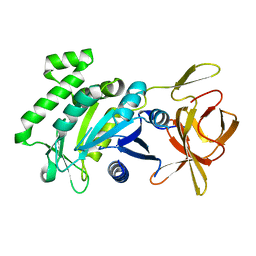 | | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6 | | Descriptor: | Elongation factor G | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6
To Be Published
|
|
5TV7
 
 | | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate. | | Descriptor: | GLUTAMINE HYDROXAMATE, Putative peptidoglycan-binding/hydrolysing protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.
To Be Published
|
|
5U47
 
 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
5UJS
 
 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
3PAJ
 
 | | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | MAGNESIUM ION, Nicotinate-nucleotide pyrophosphorylase, carboxylating | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
4NOI
 
 | | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase subunit alpha, IODIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom Crystal Structure of DNA-directed RNA Polymerase Subunit Alpha from Campylobacter jejuni.
TO BE PUBLISHED
|
|
4NEA
 
 | | 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3POL
 
 | | 2.3 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii. | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii.
TO BE PUBLISHED
|
|
3QFK
 
 | | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Krishna, S.N, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with alpha-ketoglutarate.
TO BE PUBLISHED
|
|
3QFG
 
 | | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Halavaty, A, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
