6EFE
 
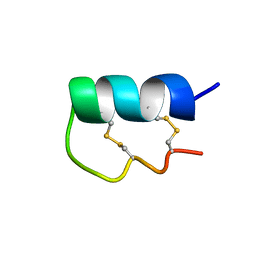 | | NMR Solution Structure of vil14a | | Descriptor: | Kappa-conotoxin vil14a | | Authors: | Dovell, S, Mari, F, Moller, C, Melaun, C. | | Deposit date: | 2018-08-16 | | Release date: | 2018-09-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Definition of the R-superfamily of conotoxins: Structural convergence of helix-loop-helix peptidic scaffolds.
Peptides, 107, 2018
|
|
2P4M
 
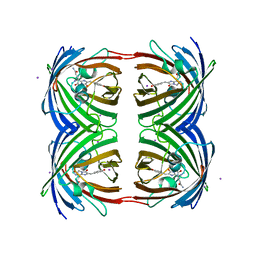 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
2ARL
 
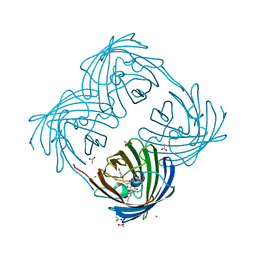 | | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding | | Descriptor: | ACETIC ACID, CHLORIDE ION, GFP-like non-fluorescent chromoprotein, ... | | Authors: | Wilmann, P.G, Battad, J, Beddoe, T, Olsen, S, Smith, S.C, Dove, S, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2005-08-19 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding
Photochem.Photobiol., 82, 2006
|
|
1MOU
 
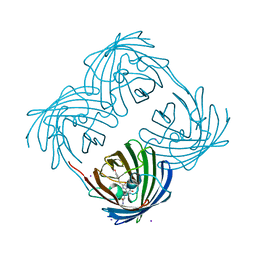 | | Crystal structure of Coral pigment | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
1MOV
 
 | | Crystal structure of Coral protein mutant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
7L7A
 
 | | Solution Structure of NuxVA | | Descriptor: | NuxVA | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2020-12-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conus venom fractions inhibit the adhesion of Plasmodium falciparum erythrocyte membrane protein 1 domains to the host vascular receptors.
J Proteomics, 234, 2020
|
|
6BX9
 
 | | Solution structure of conotoxin reg3b | | Descriptor: | Conotoxin | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural plasticity of mini-M conotoxins - expression of all mini-M subtypes by Conus regius.
FEBS J., 285, 2018
|
|
