8EDS
 
 | |
8EDR
 
 | |
8EQ0
 
 | |
8EQ1
 
 | |
8EQ3
 
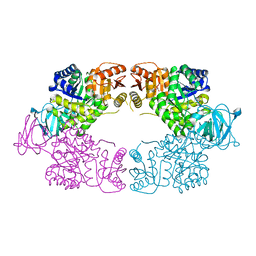 | | Escherichia coli pyruvate kinase A301T | | Descriptor: | IMIDAZOLE, MALONATE ION, Pyruvate kinase, ... | | Authors: | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | Deposit date: | 2022-10-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
8EU4
 
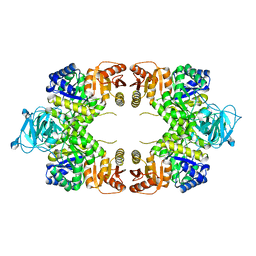 | |
4GRS
 
 | | Crystal structure of a chimeric DAH7PS | | Descriptor: | Phospho-2-dehydro-3-deoxyheptonate aldolase, 2-dehydro-3-deoxyphosphoheptonate aldolase, TYROSINE | | Authors: | Cross, P.J, Allison, T.M, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-08-26 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ON4
 
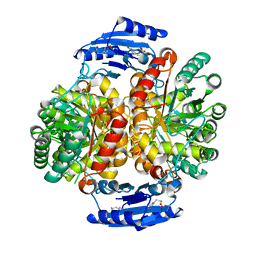
 | | Crystal structure of the GntR-type sialoregulator NanR from Escherichia coli, in complex with sialic acid | | Descriptor: | HTH-type transcriptional repressor NanR, N-acetyl-beta-neuraminic acid, ZINC ION, ... | | Authors: | Horne, C.R, Panjikar, S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the Escherichia coli sialoregulon by transcriptional repressor NanR.
J. Bacteriol., 195, 2013
|
|
6Q26
 
 | |
6Q27
 
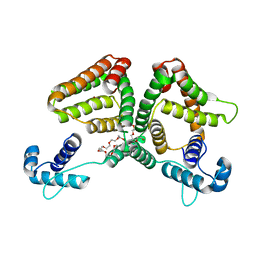
 | | N-acetylmannosamine kinase with N-acetylmannosamine from Staphylococcus aureus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, Glucokinase | | Authors: | Coombes, D, Horne, C.R, Davies, J.S, Dobson, R.C.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The basis for non-canonical ROK family function in theN-acetylmannosamine kinase from the pathogenStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6Q28
 
 | |
2OJP
 
 | | The crystal structure of a dimeric mutant of Dihydrodipicolinate synthase from E.coli- DHDPS-L197Y | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Griffin, M.D.W, Dobson, R.C.J, Antonio, L, Perugini, M.A, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-01-13 | | Release date: | 2008-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of quaternary structure in a homotetrameric enzyme.
J.Mol.Biol., 380, 2008
|
|
3WAO
 
 | | Crystal structure of Atg13 LIR-fused human LC3B_2-119 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAN
 
 | | Crystal structure of Atg13 LIR-fused human LC3A_2-121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAM
 
 | | Crystal structure of human LC3C_8-125 | | Descriptor: | CITRIC ACID, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAP
 
 | | Crystal structure of Atg13 LIR-fused human LC3C_8-125 | | Descriptor: | Autophagy-related protein 13, Microtubule-associated proteins 1A/1B light chain 3C | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3PB0
 
 | |
7MJF
 
 | | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, (4S)-4-hydroxy-2-oxoheptanedioic acid, 4-hydroxy-tetrahydrodipicolinate synthase | | Authors: | Gilkes, J, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Candidatus Liberibacter solanacearum dihydrodipicolinate synthase with pyruvate and succinic semi-aldehyde bound in active site
To Be Published
|
|
7M5I
 
 | | Endolysin from Escherichia coli O157:H7 phage FAHEc1 | | Descriptor: | Endolysin, PHOSPHATE ION | | Authors: | Love, M.J, Coombes, D, Billington, C, Dobson, R.C.J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Molecular Basis for Escherichia coli O157:H7 Phage FAHEc1 Endolysin Function and Protein Engineering to Increase Thermal Stability.
Viruses, 13, 2021
|
|
7MFS
 
 | |
7MFN
 
 | |
7MQT
 
 | |
3PB2
 
 | |
4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
