9ISP
 
 | | DUF2436 domain which is frequently found in virulence proteins from Porphyromonas gingivalis | | Descriptor: | Protease PrtH | | Authors: | Kim, B, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2024-07-18 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | First crystal structure of the DUF2436 domain of virulence proteins from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8K55
 
 | |
8K57
 
 | |
8JUO
 
 | |
8JUS
 
 | |
5BVS
 
 | | Linoleate-bound pFABP4 | | Descriptor: | Fatty acid-binding protein, LINOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
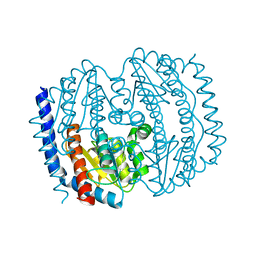
5BY2
 
 | |
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
6KRT
 
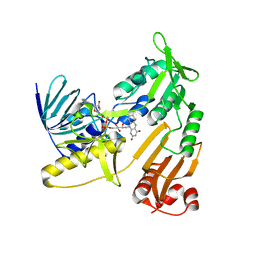 | | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, monodehydroascorbate reductase | | Authors: | Park, A.K, Do, H, Lee, J.H, Kim, H, Choi, W, Kim, I.S, Kim, H.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica
To Be Published
|
|
8WFC
 
 | |
7XRH
 
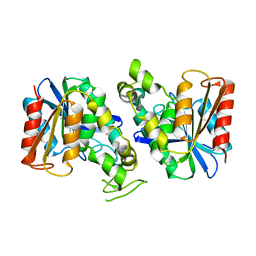 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7YVT
 
 | |
8HGU
 
 | |
8HM5
 
 | |
8ILJ
 
 | |
7CS1
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Neomycin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, NEOMYCIN | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSI
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside 2'-N-acetyltransferase, COENZYME A | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CRM
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-APO Structure | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CS0
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Paromomycin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSJ
 
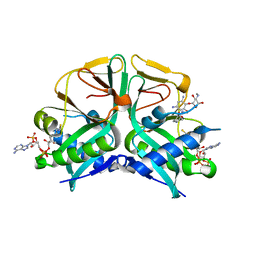 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Gentamicin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, gentamicin C1 | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7ENY
 
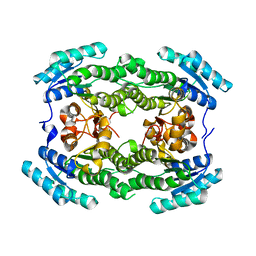 | | Crystal structure of hydroxysteroid dehydrogenase from Escherichia coli | | Descriptor: | 7alpha-hydroxysteroid dehydrogenase | | Authors: | Kim, K.-H, Lee, C.W, Pardhe, D.P, Hwang, J, Do, H, Lee, Y.M, Lee, J.H, Oh, T.-J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of an apo 7 alpha-hydroxysteroid dehydrogenase reveals key structural changes induced by substrate and co-factor binding.
J.Steroid Biochem.Mol.Biol., 212, 2021
|
|
7DVN
 
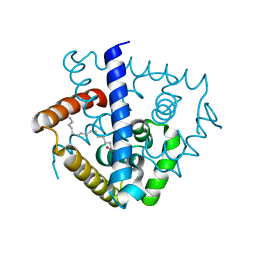 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
3UYU
 
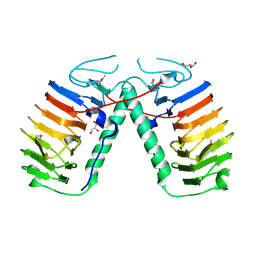 | | Structural basis for the antifreeze activity of an ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, GLYCEROL | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
