4RHE
 
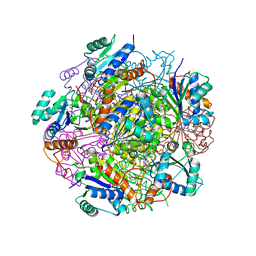 | | Crystal structure of UbiX, an aromatic acid decarboxylase from the Colwellia psychrerythraea 34H | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Do, H, Kim, S.J, Lee, C.W, Kim, H.-W, Park, H.H, Kim, H.M, Park, H, Park, H.J, Lee, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of UbiX, an aromatic acid decarboxylase from the psychrophilic bacterium Colwellia psychrerythraea that undergoes FMN-induced conformational changes.
Sci Rep, 5, 2015
|
|
4RHF
 
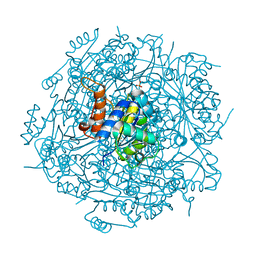 | | Crystal structure of UbiX mutant V47S from Colwellia psychrerythraea 34H | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, SULFATE ION | | Authors: | Do, H, Kim, S.J, Lee, C.W, Kim, H.-W, Park, H.H, Kim, H.M, Park, H, Park, H.J, Lee, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structure of UbiX, an aromatic acid decarboxylase from the psychrophilic bacterium Colwellia psychrerythraea that undergoes FMN-induced conformational changes.
Sci Rep, 5, 2015
|
|
8K41
 
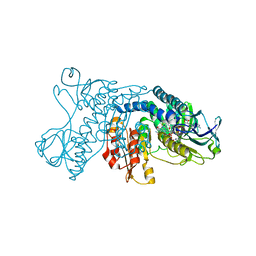 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
8K40
 
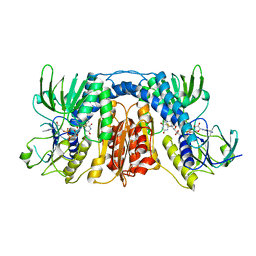 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
4ZZ7
 
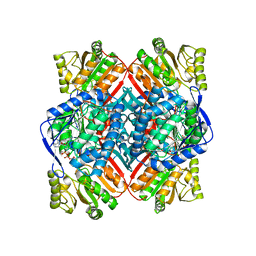 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
5D9V
 
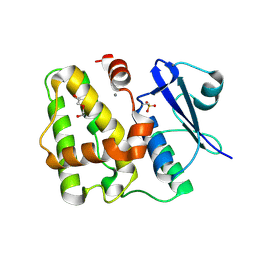 | |
5D9T
 
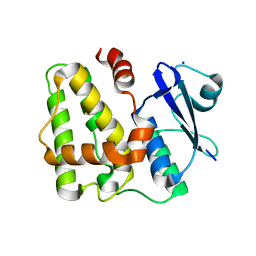 | |
5D9X
 
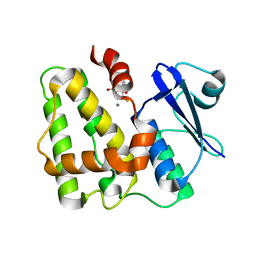 | | Dehydroascorbate reductase complexed with GSH | | Descriptor: | CALCIUM ION, Dehydroascorbate reductase, GLUTATHIONE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2015-08-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5D9W
 
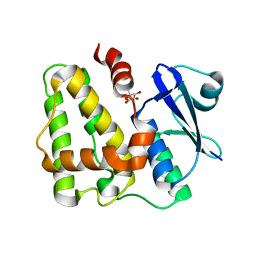 | | Dehydroascorbate reductase (OsDHAR) complexed with ASA | | Descriptor: | ASCORBIC ACID, Dehydroascorbate reductase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2015-08-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6897 Å) | | Cite: | Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
6O5C
 
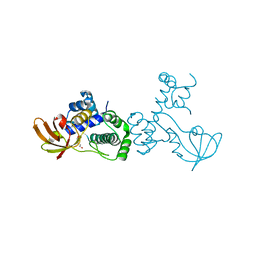 | | X-ray crystal structure of metal-dependent transcriptional regulator MtsR | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Putative metal-dependent transcriptional regulator | | Authors: | Do, H, Kumaraswami, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Metal sensing and regulation of adaptive responses to manganese limitation by MtsR is critical for group A streptococcus virulence.
Nucleic Acids Res., 47, 2019
|
|
5JLS
 
 | |
5JLU
 
 | |
6DQL
 
 | | Crystal structure of Regulator of Proteinase B RopB complexed with SIP | | Descriptor: | Regulator of Proteinase B RopB, SpeB-inducing peptide (SIP) | | Authors: | Do, H, Makthal, N, VanderWal, A.R, Olsen, R.J, Musser, J.M, Kumaraswami, M. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway.
Nat Commun, 10, 2019
|
|
4NU2
 
 | | Crystal structure of an ice-binding protein (FfIBP) from the Antarctic bacterium, Flavobacterium frigoris PS1 | | Descriptor: | Antifreeze protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NU3
 
 | | Crystal structure of mFfIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | SODIUM ION, SULFATE ION, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8GTL
 
 | | Crystal Structure of Cytochrome P450 (CYP101D5) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 CYP101D5 | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 CYP101D5 from Sphingomonas echinoides.
Int J Mol Sci, 23, 2022
|
|
7X99
 
 | |
7XJT
 
 | |
4NUH
 
 | | Crystal structure of mLeIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YC0
 
 | | Acetylesterase (LgEstI) W.T. | | Descriptor: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
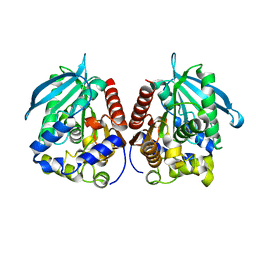 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
8HG9
 
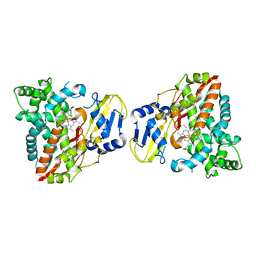 | | Cytochrome P450 steroid hydroxylase (BaCYP106A6) from Bacillus species | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 steroid hydroxylase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 Steroid Hydroxylase ( Ba CYP106A6) from Bacillus Species.
J Microbiol Biotechnol., 33, 2023
|
|
7DLS
 
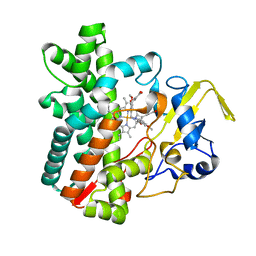 | | Cytochrome P450 (CYP105D18) complex with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Characterization of high-H 2 O 2 -tolerant bacterial cytochrome P450 CYP105D18: insights into papaverine N-oxidation.
Iucrj, 8, 2021
|
|
7DI3
 
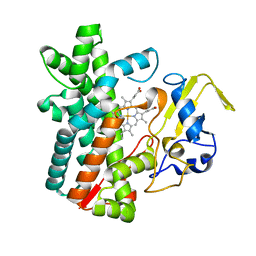 | | Cytochrome P450 (CYP105D18) W.T. | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2020-11-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Characterization of high-H 2 O 2 -tolerant bacterial cytochrome P450 CYP105D18: insights into papaverine N-oxidation.
Iucrj, 8, 2021
|
|
7EHK
 
 | | Crystal structure of C107S mutant of FfIBP | | Descriptor: | CHLORIDE ION, Ice-binding protein | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2021-03-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of rigidity of ice-binding protein (FfIBP) for hyperthermal hysteresis activity and microbial survival.
Int.J.Biol.Macromol., 204, 2022
|
|
