1K5Q
 
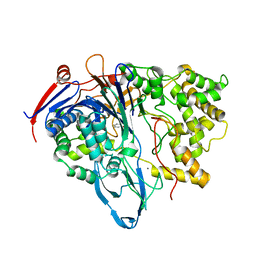 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PAA | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
4BAA
 
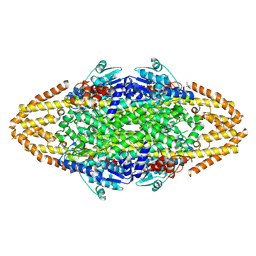 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
1KCK
 
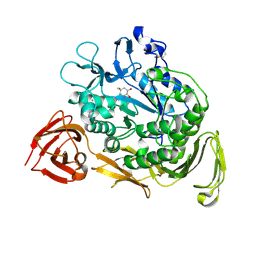 | | Bacillus circulans strain 251 Cyclodextrin glycosyl transferase mutant N193G | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
1KCL
 
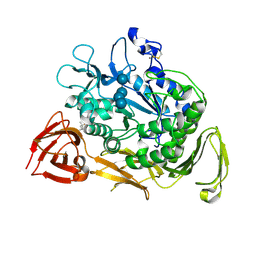 | | Bacillus ciruclans strain 251 Cyclodextrin glycosyl transferase mutant G179L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cyclodextrin glycosyltransferase, ... | | Authors: | Rozeboom, H.J, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The remote substrate binding subsite -6 in cyclodextrin-glycosyltransferase controls the transferase activity of the enzyme via an induced-fit mechanism.
J.Biol.Chem., 277, 2002
|
|
4CVB
 
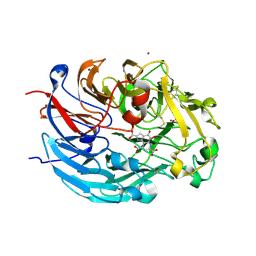 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes | | Descriptor: | ALCOHOL DEHYDROGENASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
4AO9
 
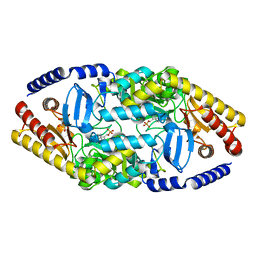 | | Biochemical properties and crystal structure of a novel beta- phenylalanine aminotransferase from Variovorax paradoxus | | Descriptor: | BETA-PHENYLALANINE AMINOTRANSFERASE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Crismaru, C.G, Wybenga, G.G, Szymanski, W, Wijma, H.J, Wu, B, deWildeman, S, Poelarends, G.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-03-25 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical Properties and Crystal Structure of a Novel Beta-Phenylalanine Aminotransferase from Variovorax Paradoxus
Appl.Environ.Microbiol., 79, 2013
|
|
4AMW
 
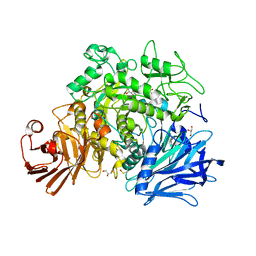 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-idosyl- fluoride | | Descriptor: | 5-fluoro-alpha-L-idopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4AMC
 
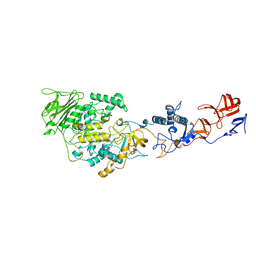 | | Crystal structure of Lactobacillus reuteri 121 N-terminally truncated glucansucrase GTFA | | Descriptor: | CALCIUM ION, GLUCANSUCRASE | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-03-08 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Alpha-1,6/Alpha-1,4-Specific Glucansucrase Gtfa from Lactobacillus Reuteri 121
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4CVC
 
 | | Crystal structure of quinone-dependent alcohol dehydrogenase from Pseudogluconobacter saccharoketogenenes with zinc in the active site | | Descriptor: | ALCOHOL DEHYDROGENASE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Yu, S, Mikkelsen, R, Nikolaev, I, Mulder, H, Dijkstra, B.W. | | Deposit date: | 2014-03-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Quinone-Dependent Alcohol Dehydrogenase from Pseudogluconobacter Saccharoketogenes. A Versatile Dehydrogenase Oxidizing Alcohols and Carbohydrates.
Protein Sci., 24, 2015
|
|
1I6W
 
 | | THE CRYSTAL STRUCTURE OF BACILLUS SUBTILIS LIPASE: A MINIMAL ALPHA/BETA HYDROLASE ENZYME | | Descriptor: | CADMIUM ION, LIPASE A | | Authors: | van Pouderoyen, G, Eggert, T, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2001-03-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme.
J.Mol.Biol., 309, 2001
|
|
1LG1
 
 | |
1LG2
 
 | |
4AT0
 
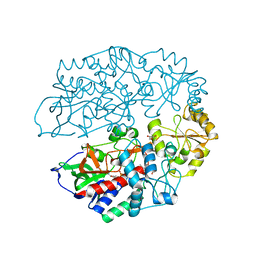 | | The crystal structure of 3-ketosteroid-delta4-(5alpha)-dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | 3-KETOSTEROID-DELTA4-5ALPHA-DEHYDROGENASE, ACETATE ION, CHLORIDE ION, ... | | Authors: | van Oosterwijk, N, Knol, J, Dijkhuizen, L, van der Geize, R, Dijkstra, B.W. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Catalytic Mechanism of 3-Ketosteroid-{Delta}4-(5Alpha)-Dehydrogenase from Rhodococcus Jostii Rha1 Genome.
J.Biol.Chem., 287, 2012
|
|
1K5S
 
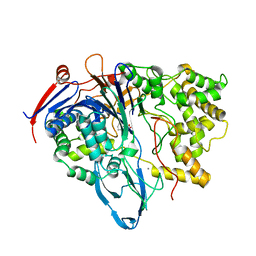 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PPA | | Descriptor: | CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, PENICILLIN G ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1LTM
 
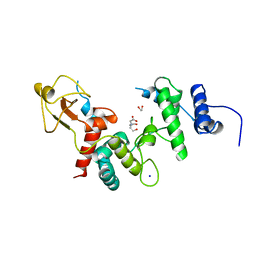 | | ACCELERATED X-RAY STRUCTURE ELUCIDATION OF A 36 KDA MURAMIDASE/TRANSGLYCOSYLASE USING WARP | | Descriptor: | 1,2-ETHANEDIOL, 36 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE, BICINE, ... | | Authors: | Van Asselt, E.J, Dijkstra, B.W. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1LQ0
 
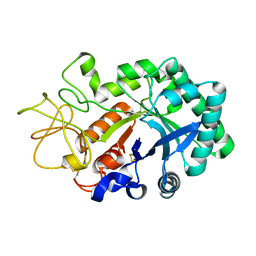 | |
