2P9E
 
 | |
7MPB
 
 | | SARS Coronavirus-2 Main Protease 3CL-pro binding Ascorbate | | Descriptor: | 3C-like proteinase, ASCORBIC ACID, TRIFLUOROETHANOL | | Authors: | Pandey, S, Malla, T.N, Stojkovic, E.A, Schmidt, M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vitamin C inhibits SARS coronavirus-2 main protease essential for viral replication
Biorxiv, 2021
|
|
4RZJ
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with N-acetylglucosamine at 1.98 Angstrom resolution using crystals grown in different conditions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Pandey, S, Kushwaha, G.S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with N-acetylglucosamine at 1.98 Angstrom resolution using crystals grown in different conditions
TO BE PUBLISHED
|
|
2LY3
 
 | |
6P5G
 
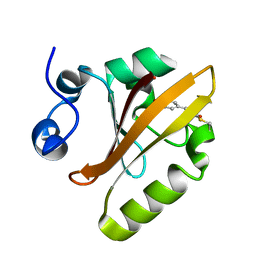 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P4I
 
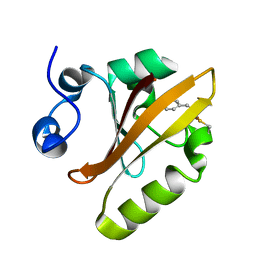 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
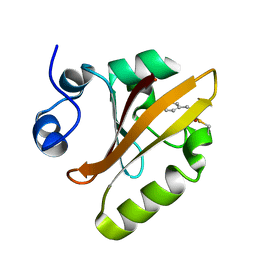 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5D
 
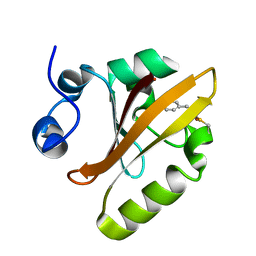 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6PU2
 
 | | Dark, Mutant H275T , 100K, PCM Myxobacterial Phytochrome, P2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6PTQ
 
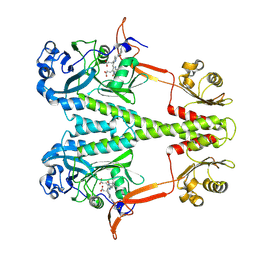 | | Dark, Room Temperature, PCM Myxobacterial Phytochrome, P2, Wild Type | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6PTX
 
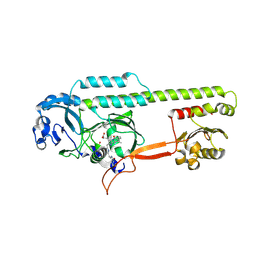 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
1RMJ
 
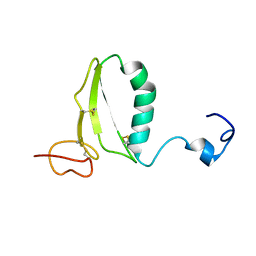 | | C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II | | Descriptor: | Insulin-like growth factor binding protein 6 | | Authors: | Headey, S.J, Keizer, D.W, Yao, S, Brasier, G, Kantharidis, P, Bach, L.A, Norton, R.S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II.
Mol.Endocrinol., 18, 2004
|
|
7FHW
 
 | |
2MHC
 
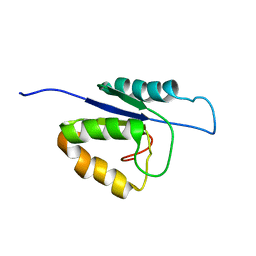 | | NMR structure of the catalytic domain of the large serine resolvase TnpX | | Descriptor: | TnpX | | Authors: | Headey, S.J, Sivakumaran, A, Adams, V, Rodgers, A.J.W, Rood, J.I, Scanlon, M.J, Wilce, M.C.J. | | Deposit date: | 2013-11-20 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA Binding of the Catalytic of the Large Serine Resolvase Tnpx
To be Published
|
|
5DVF
 
 | | Crystal structure of unliganded periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, SULFATE ION | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
5DVJ
 
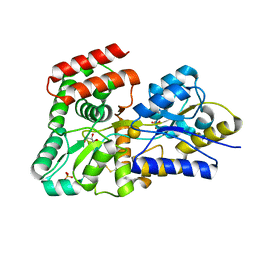 | | Crystal structure of galactose complexed periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, GLYCEROL, SULFATE ION, ... | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
7FHU
 
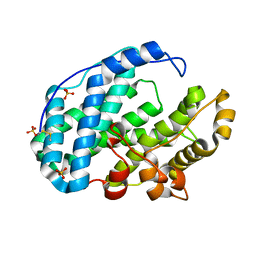 | |
7FHV
 
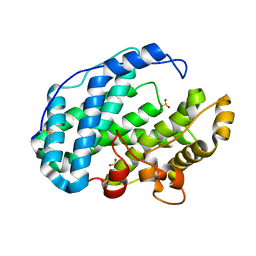 | |
7FHZ
 
 | |
7FI1
 
 | |
7FI2
 
 | |
7FI0
 
 | | Crystal structure of Multi-functional Polysaccharide lyase Smlt1473 (WT) from Stenotrophomonas maltophilia (strain K279a) in ManA bound form at pH-5.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polysaccharide lyase, SULFATE ION, ... | | Authors: | Pandey, S, Berger, B.W, Acharya, R. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural insights into the mechanism of pH-selective substrate specificity of the polysaccharide lyase Smlt1473.
J.Biol.Chem., 297, 2021
|
|
7FHY
 
 | |
7FHX
 
 | |
