6ZIF
 
 | |
6WKT
 
 | |
5ARM
 
 | | APO-CSP3 (COPPER STORAGE PROTEIN 3) FROM METHYLOSINUS | | Descriptor: | CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.194 Å) | | Cite: | Novel Cytosolic Copper Storage Proteins
To be Published
|
|
4OZ7
 
 | |
5ARN
 
 | | Cu(I)-CSP3 (COPPER STORAGE PROTEIN 3) FROM METHYLOSINUS | | Descriptor: | COPPER (I) ION, CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Cytosolic Copper Storage Proteins
To be Published
|
|
1X9U
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the reduced form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
5NQO
 
 | |
5NQN
 
 | |
5NQM
 
 | |
1X9R
 
 | | Umecyanin from Horse Raddish- Crystal Structure of the oxidised form | | Descriptor: | COPPER (II) ION, Umecyanin | | Authors: | Koch, M, Velarde, M, Harrison, M.D, Echt, S, Fischer, M, Messerschmidt, A, Dennison, C. | | Deposit date: | 2004-08-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Oxidized and Reduced Stellacyanin from Horseradish Roots
J.Am.Chem.Soc., 127, 2005
|
|
5FIG
 
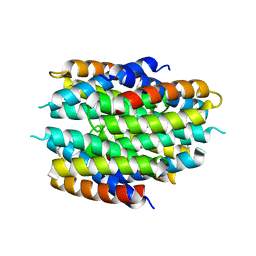 | | APO-CSP3 (COPPER STORAGE PROTEIN 3) FROM BACILLUS SUBTILIS | | Descriptor: | CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity.
Sci Rep, 6, 2016
|
|
5FJD
 
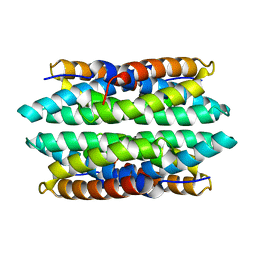 | | APO-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER STORAGE PROTEIN 1 | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
5FJE
 
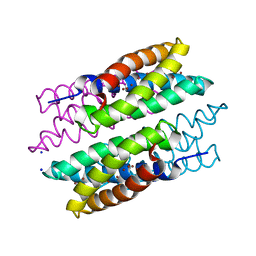 | | CU(I)-CSP1 (COPPER STORAGE PROTEIN 1) FROM METHYLOSINUS TRICHOSPORIUM OB3B | | Descriptor: | COPPER (I) ION, COPPER STORAGE PROTEIN 1, SODIUM ION | | Authors: | Vita, N, Platsaki, S, Basle, A, Allen, S.J, Paterson, N.G, Crombie, A.T, Murrell, J.C, Waldron, K.J, Dennison, C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Four-Helix Bundle Stores Copper for Methane Oxidation.
Nature, 525, 2015
|
|
3CVB
 
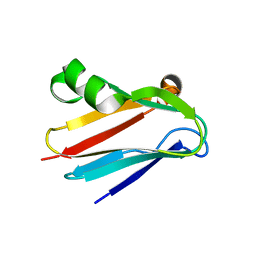 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (I) ION, Plastocyanin | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
3CVC
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (II) ION, MAGNESIUM ION, Plastocyanin | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
3CVD
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (I) ION, Plastocyanin, ZINC ION | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
2VP1
 
 | | Fe-FutA2 from Synechocystis PCC6803 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE (III) ION, ... | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
2UX7
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of loop shortening on the metal binding site of cupredoxin pseudoazurin.
Biochemistry, 46, 2007
|
|
2UX6
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXF
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXG
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2VOZ
 
 | | Apo FutA2 from Synechocystis PCC6803 | | Descriptor: | PERIPLASMIC IRON-BINDING PROTEIN, SULFATE ION | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
2GHZ
 
 | |
2GI0
 
 | |
2JSC
 
 | | NMR structure of the cadmium metal-sensor CMTR from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Transcriptional regulator Rv1994c/MT2050 | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Cavet, J.S, Dennison, C, Graham, A.I, Harvie, D.R, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Analysis of Cadmium Sensing by Winged Helix Repressor CmtR.
J.Biol.Chem., 282, 2007
|
|
