4DR7
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DR1
 
 | | Crystal structure of the apo 30S ribosomal subunit from Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DV3
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, C912A, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DR6
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DUZ
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
3FUU
 
 | | T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in complex with Adenosine in space group P212121 | | Descriptor: | ADENOSINE, Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUT
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P21212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUW
 
 | | T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in complex with 5'-methylthioadenosine in space group P212121 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUV
 
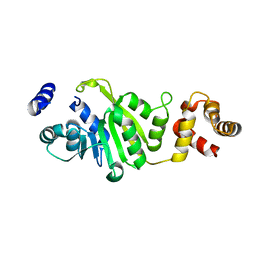 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P43212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3G8B
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group I222 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
4DV1
 
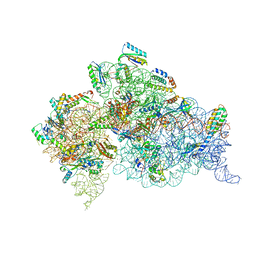 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G, bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.849 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DV4
 
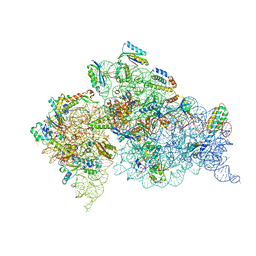 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A914G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DV6
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, A915G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.297 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DV0
 
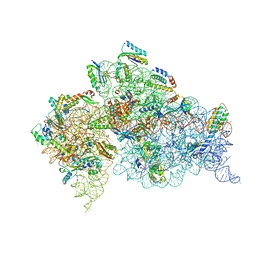 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U20G | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.853 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4DUY
 
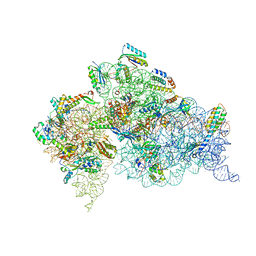 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C | | Descriptor: | 16S rRNA, MAGNESIUM ION, ZINC ION, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3.386 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
7E1J
 
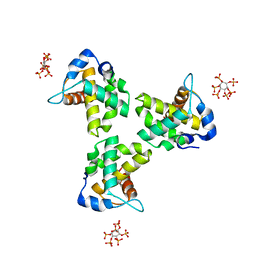 | |
7EK8
 
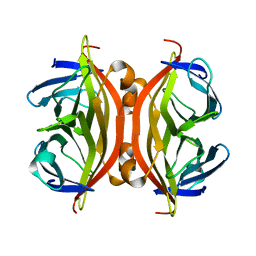 | |
7EK9
 
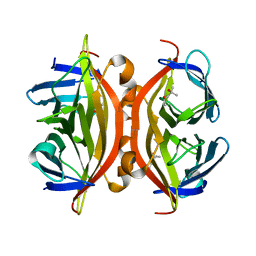 | | Crystal structure of apo streptavidin at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | DeMirci, H, Ayan, E, Destan, E, Ertem, F.B. | | Deposit date: | 2021-04-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of apo streptavidin at cryogenic temperature
To Be Published
|
|
7EUM
 
 | |
7E1K
 
 | |
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
7YKN
 
 | | Crystal structure of (6-4) photolyase from Vibrio cholerae | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cakilkaya, B, Kavakli, I.H, DeMirci, H. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Vibrio cholerae (6-4) photolyase reveals interactions with cofactors and a DNA-binding region.
J.Biol.Chem., 299, 2023
|
|
8IVJ
 
 | |
6DTI
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an unmodifed anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-06-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
8IWI
 
 | |
