6YN2
 
 | |
6TY7
 
 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
1MJ5
 
 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
2BFN
 
 | | The crystal structure of the complex of the haloalkane dehalogenase LinB with the product of dehalogenation reaction 1,2-dichloropropane. | | Descriptor: | (2S)-2,3-DICHLOROPROPAN-1-OL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Banas, P, Otyepka, M, Jerabek, P, Vevodova, J, Bohac, M, Damborsky, J. | | Deposit date: | 2004-12-09 | | Release date: | 2006-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Weak Activity of Haloalkane Dehalogenase Linb with 1,2,3-Trichloropropane Revealed by X-Ray Crystallography and Microcalorimetry
Appl.Environ.Microbiol., 73, 2007
|
|
3SK0
 
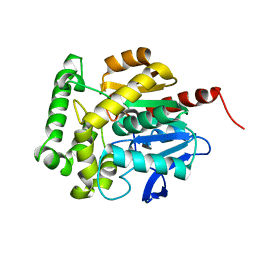 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
6F9O
 
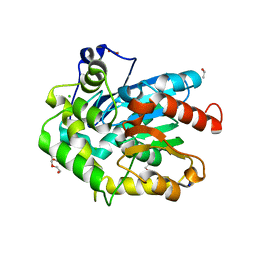 | | Crystal structure of cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Haloalkane dehalogenase, ... | | Authors: | Tratsiak, K, Prudnikova, T, Drienovska, I, Damborsky, J, Brynda, J, Pachl, P, Kuty, M, Chaloupkova, R, Kuta Smatanova, I, Rezacova, P. | | Deposit date: | 2017-12-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5FLK
 
 | |
5LKA
 
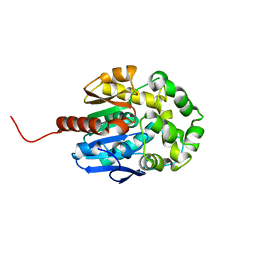 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
2V9Z
 
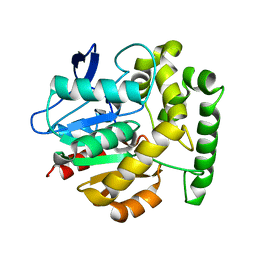 | | Structure of the Rhodococcus haloalkane dehalogenase mutant with enhanced enantioselectivity | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Koudelakova, T, Prokop, Z, Sato, Y, Lapkouski, M, Chovancova, E, Monincova, M, Jesenska, A, Emmer, J, Senda, T, Nagata, Y, Kuta Smatanova, I, Damborsky, J. | | Deposit date: | 2007-08-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational Engineering of Rhodococcus Haloalkane Dehalogenase with Enhanced Enantioselectivity
To be Published
|
|
4K2A
 
 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RK4
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6G75
 
 | | Crystal structure of the common ancestor of haloalkane dehalogenases and Renilla luciferase (AncHLD-RLuc) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Common ancestor of haloalkane dehalogenase and Renilla luciferase (AncHLD-RLuc), ... | | Authors: | Chaloupkova, R, Waterman, J, Marek, M, Damborsky, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Light-Emitting Dehalogenases: Reconstruction of Multifunctional Biocatalysts
Acs Catalysis, 2019
|
|
4WDQ
 
 | | Crystal structure of haloalkane dehalogenase LinB32 mutant (L177W) from Sphingobium japonicum UT26 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, MAGNESIUM ION | | Authors: | Degtjarik, O, Rezacova, P, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WDR
 
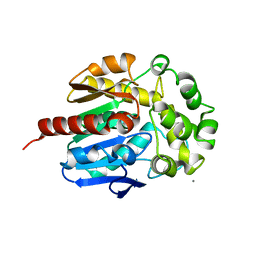 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
1G4H
 
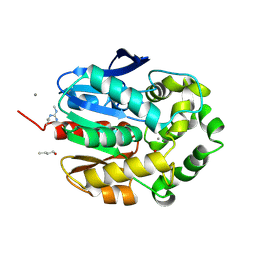 | | LINB COMPLEXED WITH BUTAN-1-OL | | Descriptor: | 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, 1-BUTANOL, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
4E46
 
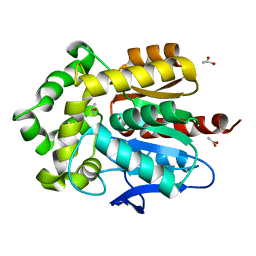 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA in complex with 2-propanol | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Stsiapanava, A, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents.
Chembiochem, 14, 2013
|
|
1G5F
 
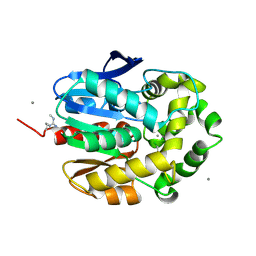 | | STRUCTURE OF LINB COMPLEXED WITH 1,2-DICHLOROETHANE | | Descriptor: | 1,2-DICHLOROETHANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
1G42
 
 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | Descriptor: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-10-26 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
6S6E
 
 | |
6S97
 
 | |
1CV2
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 AT 1.6 A resolution | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-08-22 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
6SP8
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
1D07
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
6SP5
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6XTC
 
 | |
