6XT8
 
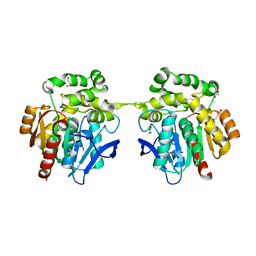 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
1K63
 
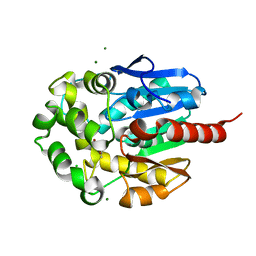 | | Complex of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis with UT26 2-BROMO-2-PROPENE-1-OL at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, 2-BROMO-2-PROPENE-1-OL, BROMIDE ION, ... | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1K6E
 
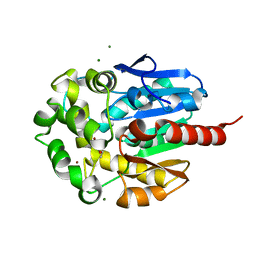 | | COMPLEX OF HYDROLYTIC HALOALKANE DEHALOGENASE LINB FROM SPHINGOMONAS PAUCIMOBILIS UT26 WITH 1,2-PROPANEDIOL (PRODUCT OF DEHALOGENATION OF 1,2-DIBROMOPROPANE) AT 1.85A | | Descriptor: | 1-BROMOPROPANE-2-OL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Streltsov, V.A, Prokop, Z, Damborsky, J, Nagata, Y, Oakley, A, Wilce, M.C.J. | | Deposit date: | 2001-10-16 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates.
Biochemistry, 42, 2003
|
|
1K5P
 
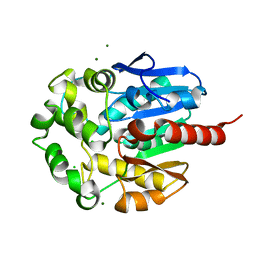 | | Hydrolytic haloalkane dehalogenase LINB from sphingomonas paucimobilis UT26 at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
6Y9E
 
 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
6Y9G
 
 | |
6XY9
 
 | | Crystal structure of haloalkane dehalogenase DbeA-M1 loop variant from Bradyrhizobium elkanii | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Rezacova, P, Kuta Smatanova, I, Chaloupkova, R, Damborsky, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and catalytic effects of surface loop-helix transplantation within haloalkane dehalogenase family.
Comput Struct Biotechnol J, 18, 2020
|
|
6Y9F
 
 | |
