3HSF
 
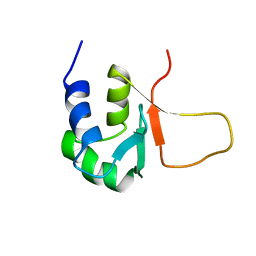 | | HEAT SHOCK TRANSCRIPTION FACTOR (HSF) | | Descriptor: | HEAT SHOCK TRANSCRIPTION FACTOR | | Authors: | Damberger, F.F, Pelton, J.G, Liu, C, Cho, H, Harrison, C.J, Nelson, H.C.M, Wemmer, D.E. | | Deposit date: | 1995-08-07 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and dynamics of the DNA-binding domain of the heat shock factor from Kluyveromyces lactis.
J.Mol.Biol., 254, 1995
|
|
2JPO
 
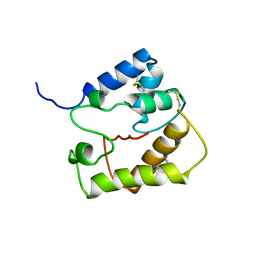 | |
2L52
 
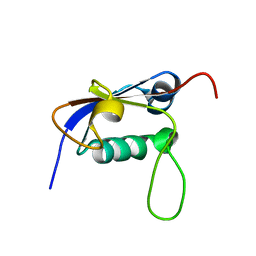 | | Solution structure of the small archaeal modifier protein 1 (SAMP1) from Methanosarcina acetivorans | | Descriptor: | METHANOSARCINA ACETIVORANS SAMP1 HOMOLOG | | Authors: | Damberger, F.F, Ranjan, N, Sutter, M, Allain, F.H.-T, Weber-Ban, E. | | Deposit date: | 2010-10-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins.
J.Mol.Biol., 405, 2011
|
|
5M8I
 
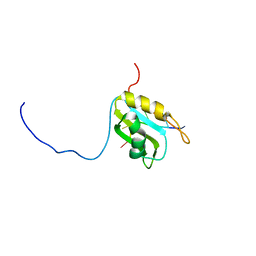 | |
1XFR
 
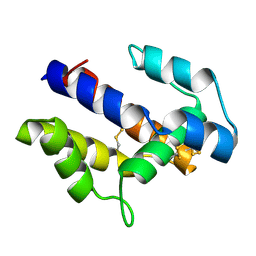 | |
6G04
 
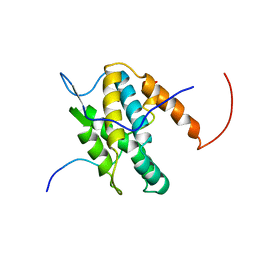 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6G03
 
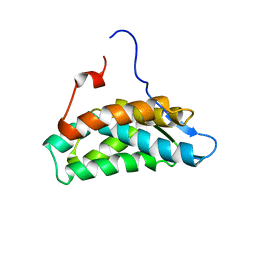 | | NMR Solution Structure of yeast TSR2(1-152) | | Descriptor: | Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
4BS2
 
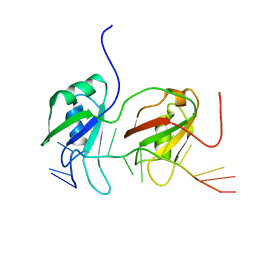 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B0R
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway | | Descriptor: | DEAMIDASE-DEPUPYLASE DOP | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway.
Nat Commun, 3, 2012
|
|
4B0T
 
 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4B0S
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
2KFL
 
 | |
2KFM
 
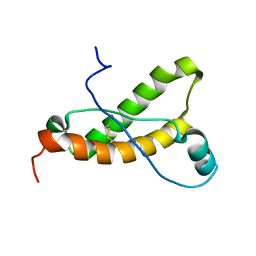 | |
2KFO
 
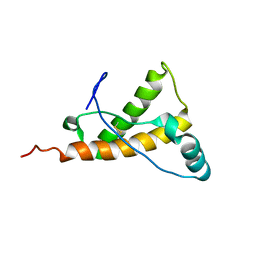 | |
2L1K
 
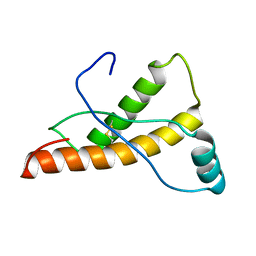 | | Mouse prion protein (121-231) containing the substitutions Y169A, Y225A, and Y226A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Temperature-dependent conformational exchange in the cellular form of prion proteins
To be Published
|
|
2LFW
 
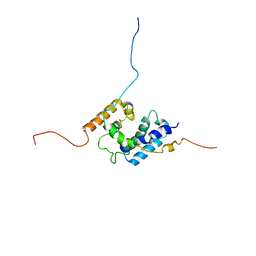 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L40
 
 | | Mouse prion protein (121-231) containing the substitution Y169A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L39
 
 | | Mouse prion protein fragment 121-231 AT 37 C | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2N3O
 
 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
2L1E
 
 | | Mouse prion protein (121-231) containing the substitution F175A | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2012-10-24 | | Method: | SOLUTION NMR | | Cite: | Prion Protein mPrP[F175A](121-231): Structure and Stability in Solution.
J.Mol.Biol., 423, 2012
|
|
2L1D
 
 | | Mouse prion protein (121-231) containing the substitution Y169G | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2011-11-09 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L1H
 
 | |
2KU6
 
 | |
2MXY
 
 | | Solution structure of hnRNP C RRM in complex with 5'-AUUUUUC-3' RNA | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
2MZ1
 
 | | Solution structure of hnRNP C RRM in complex with 5'-UUUUC-3' RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
