1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
1QH1
 
 | | NITROGENASE MOFE PROTEIN FROM KLEBSIELLA PNEUMONIAE, PHENOSAFRANIN OXIDIZED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-10 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
2FJ9
 
 | | High resolution crystal structure of the unliganded human ACBP | | Descriptor: | Acyl-CoA-binding protein, CHLORIDE ION, LEAD (II) ION, ... | | Authors: | Taskinen, J.P, van Aalten, D.M, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2006-01-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of unliganded and liganded human liver ACBP reveal a new mode of binding for the acyl-CoA ligand.
Proteins, 66, 2006
|
|
1QLK
 
 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
1PR3
 
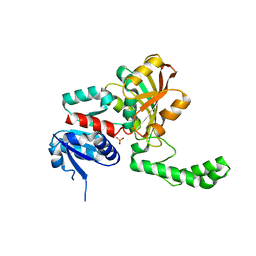 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2CG0
 
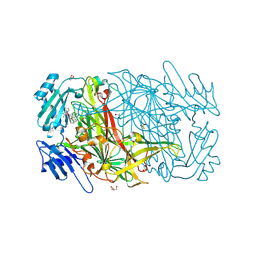 | | AGAO in complex with wc9a (Ru-wire inhibitor, 9-carbon linker, data set a) | | Descriptor: | COPPER (II) ION, GLYCEROL, PHENYLETHYLAMINE OXIDASE, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2006-02-27 | | Release date: | 2007-05-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enantiomer-Specific Binding of Ruthenium(II) Molecular Wires by the Amine Oxidase of Arthrobacter Globiformis.
J.Am.Chem.Soc., 130, 2008
|
|
1N8C
 
 | | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex | | Descriptor: | (9S,10R)-9-HYDROXY-7,8,9,10-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Volk, D.E, Thiviyanathan, V, Rice, J.S, Luxon, B.A, Shah, J.H, Yagi, H, Sayer, J.M, Yeh, H.J.C, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex
Biochemistry, 42, 2003
|
|
1NG7
 
 | |
2DAA
 
 | |
1QGU
 
 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, DITHIONITE-REDUCED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-06 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
2FIK
 
 | | Structure of a microbial glycosphingolipid bound to mouse CD1d | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, D, Zajonc, D.M. | | Deposit date: | 2005-12-29 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6L4M
 
 | | Crystal structure of vicilin from Solanum lycopersicum (tomato) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Vicilin | | Authors: | Jain, A, Salunke, D.M. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Comparative study of 7S globulin from Corylus avellana and Solanum lycopersicum revealed importance of salicylic acid and Cu-binding loop in modulating their function.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6L7N
 
 | |
6L8B
 
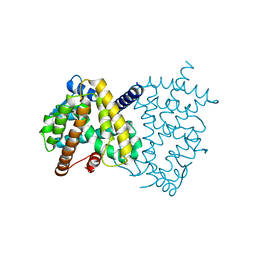 | | The ligand-free structure of human PPARgamma LBD | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, D.M, Han, B.W. | | Deposit date: | 2019-11-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Cyclin-Dependent Kinase 5 Inhibitor Butyrolactone I Elicits a Partial Agonist Activity of Peroxisome Proliferator-Activated Receptor gamma.
Biomolecules, 10, 2020
|
|
4UUC
 
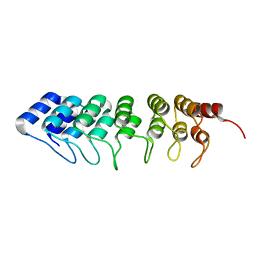 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
4UYI
 
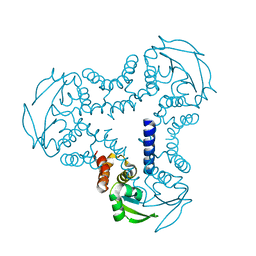 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
4WUB
 
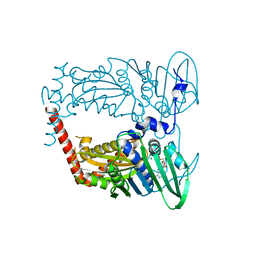 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WUC
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WJB
 
 | | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Putative amidohydrolase/peptidase, SULFATE ION, ... | | Authors: | Lukacs, C.M, Dranow, D.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia
To Be Published
|
|
4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
7K5V
 
 | | OXA-48 bound by Compound 3.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-09-17 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
