1YTE
 
 | |
1YQ6
 
 | | PRD1 vertex protein P5 | | Descriptor: | Minor capsid protein | | Authors: | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
2DD1
 
 | | Three consecutive sheared GA pairs in 5'GGUGGAGGCU/3'PCCAAAGCCG | | Descriptor: | 5'-R(*GP*CP*CP*GP*AP*AP*AP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*GP*AP*GP*GP*CP*U)-3' | | Authors: | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | Deposit date: | 2006-01-19 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
1YTD
 
 | |
3R4Y
 
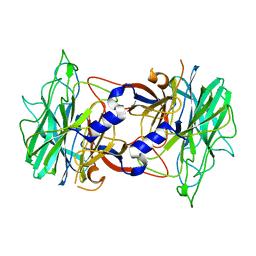 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
1YFV
 
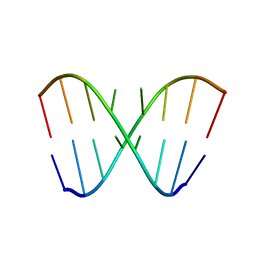 | |
2AXV
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2DD3
 
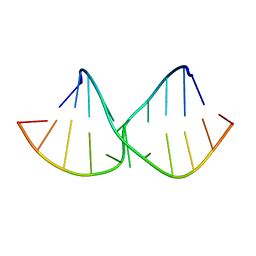 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: II. The minor conformation with A6/A5/A16 stack | | Descriptor: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | Authors: | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | Deposit date: | 2006-01-19 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2DD2
 
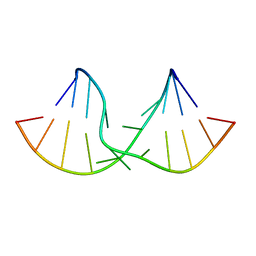 | | An alternating sheared AA pair in 5'GGUGAAGGCU/3'PCCGAAGCCG: I. The major conformation with A6/A15/A16 stack | | Descriptor: | 5'-R(*GP*CP*CP*GP*AP*AP*GP*CP*CP*(P5P))-3', 5'-R(*GP*GP*UP*GP*AP*AP*GP*GP*CP*U)-3' | | Authors: | Chen, G, Kennedy, S.D, Krugh, T.R, Turner, D.H. | | Deposit date: | 2006-01-19 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Alternating Sheared AA Pair and Elements of Stability for a Single Sheared Purine-Purine Pair Flanked by Sheared GA Pairs in RNA
Biochemistry, 45, 2006
|
|
2BA2
 
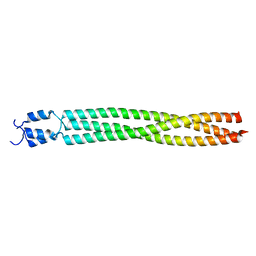 | | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae | | Descriptor: | Hypothetical UPF0134 protein MPN010 | | Authors: | Shin, D.H, Kim, J.-S, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-10-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae.
Protein Sci., 15, 2006
|
|
2ADG
 
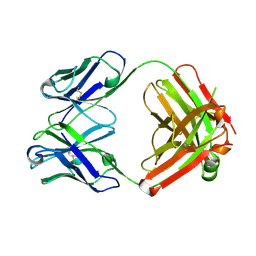 | | Crystal structure of monoclonal anti-CD4 antibody Q425 | | Descriptor: | Q425 Fab Light chain, Q425 Fab heavy chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2DEN
 
 | |
2KY1
 
 | |
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2KY2
 
 | |
2KXZ
 
 | |
1NEH
 
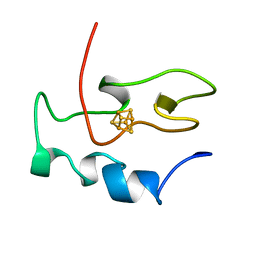 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1NK3
 
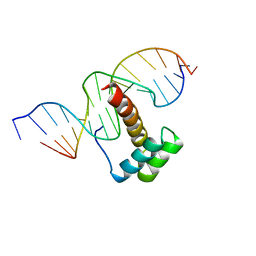 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK2
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
2L8F
 
 | |
1IK4
 
 | | X-ray Structure of Methylglyoxal Synthase from E. coli Complexed with Phosphoglycolohydroxamic Acid | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHOGLYCOLOHYDROXAMIC ACID | | Authors: | Marks, G.T, Harris, T.K, Massiah, M.A, Mildvan, A.S, Harrison, D.H.T. | | Deposit date: | 2001-05-02 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic implications of methylglyoxal synthase complexed with phosphoglycolohydroxamic acid as observed by X-ray crystallography and NMR spectroscopy.
Biochemistry, 40, 2001
|
|
2LIU
 
 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
2MH2
 
 | | Structural insights into the DNA recognition and protein interaction domains reveal fundamental homologous DNA pairing properties of HOP2 | | Descriptor: | Homologous-pairing protein 2 homolog | | Authors: | Moktan, H, Guiraldelli, M.F, Eyter, C.A, Zhao, W, Camerini-Otero, R.D, Sung, P, Zhou, D.H, Pezza, R.J. | | Deposit date: | 2013-11-13 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Properties of the Winged Helix Domain of the Meiotic Recombination HOP2 Protein.
J.Biol.Chem., 289, 2014
|
|
2MIZ
 
 | |
1HRQ
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
