1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
2EZT
 
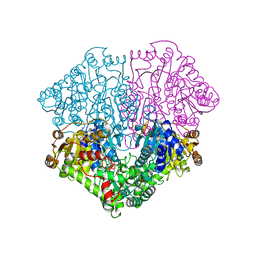 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2DKA
 
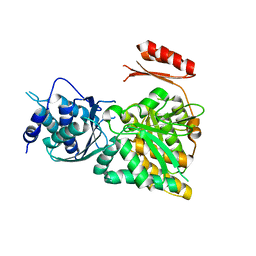 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DSM
 
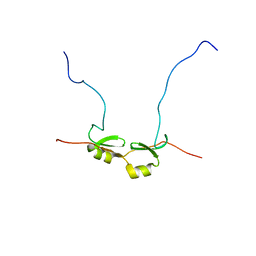 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
2DWM
 
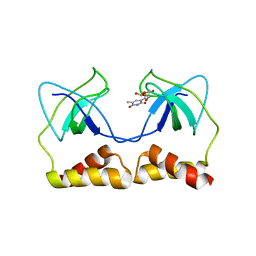 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2E35
 
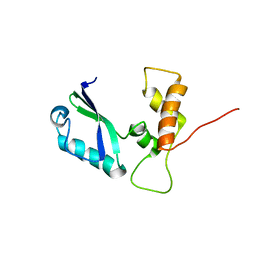 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
3PDF
 
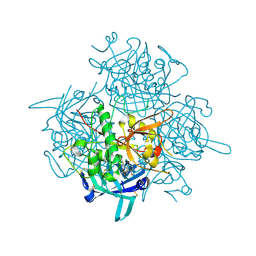 | | Discovery of Novel Cyanamide-Based Inhibitors of Cathepsin C | | Descriptor: | 2,5-dibromo-N-{(3R,5S)-1-[(Z)-iminomethyl]-5-methylpyrrolidin-3-yl}benzenesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, B, Laine, D. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of novel cyanamide-based inhibitors of cathepsin C.
Acs Med.Chem.Lett., 2, 2011
|
|
1M9U
 
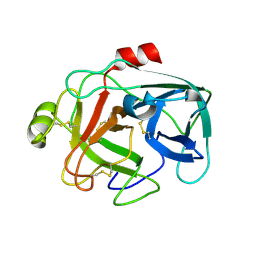 | |
2ERX
 
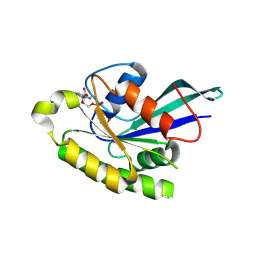 | | Crystal Structure of DiRas2 in Complex With GDP and Inorganic Phosphate | | Descriptor: | GTP-binding protein Di-Ras2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Papagrigoriou, E, Yang, X, Elkins, J, Niesen, F.E, Burgess, N, Salah, E, Fedorov, O, Ball, L.J, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of DiRas2
To be Published
|
|
2F2B
 
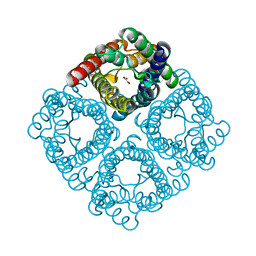 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2F3V
 
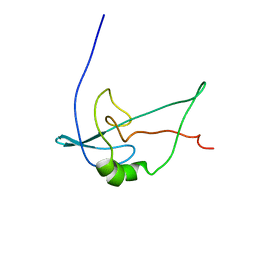 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2FH4
 
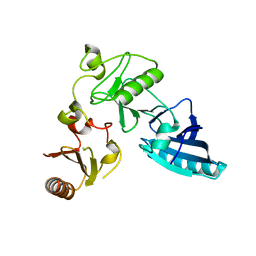 | | C-terminal half of gelsolin soaked in EGTA at pH 8 | | Descriptor: | Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
2F5Y
 
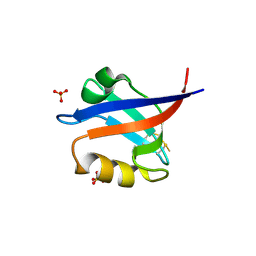 | | Crystal Structure of the PDZ Domain from Human RGS-3 | | Descriptor: | SULFATE ION, regulator of G-protein signalling 3 isoform 1 | | Authors: | Ugochukwu, E, Berridge, G, Johansson, C, Smee, C, Savitsky, P, Burgess, N, Colebrook, S, Yang, X, Elkins, J, Doyle, D, Turnbull, A, Papagrigoriou, E, Debreczeni, J, Bunkoczi, G, Gorrec, F, von Delft, F, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the PDZ Domain from Human RGS-3
To be Published
|
|
2FIK
 
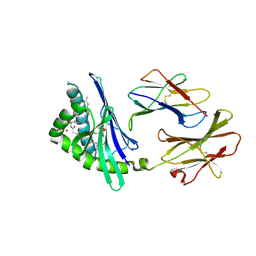 | | Structure of a microbial glycosphingolipid bound to mouse CD1d | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, D, Zajonc, D.M. | | Deposit date: | 2005-12-29 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FE5
 
 | | The Crystal Structure of the Second PDZ Domain of Human DLG3 | | Descriptor: | GLYCEROL, Presynaptic protein SAP102, SULFATE ION | | Authors: | Ugochukwu, E, Phillips, C, Schoch, G, Berridge, G, Salah, E, Colebrook, S, Smee, C, Savitsky, P, Bray, J, Elkins, J, Doyle, D, Bunkoczi, G, Debreczeni, J, Turnbull, A, Gorrec, F, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-15 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Crystal Structure of the Second PDZ Domain of Human DLG3
To be Published
|
|
1MMC
 
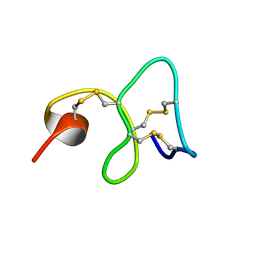 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1MQ7
 
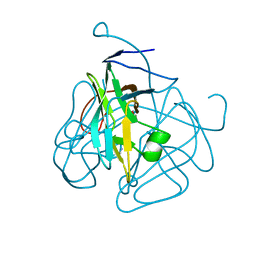 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1M4R
 
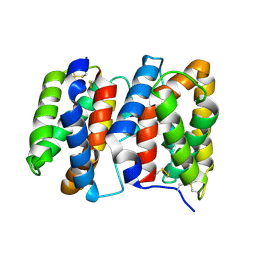 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN INTERLEUKIN-22 | | Descriptor: | Interleukin-22 | | Authors: | Nagem, R.A.P, Colau, D, Dumoutier, L, Renauld, J.-C, Ogata, C, Polikarpov, I. | | Deposit date: | 2002-07-03 | | Release date: | 2003-07-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Recombinant Human Interleukin-22
Structure, 10, 2002
|
|
1LVH
 
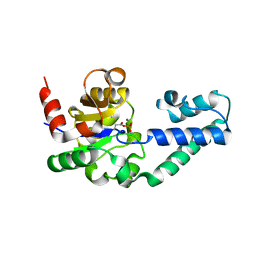 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
2EZU
 
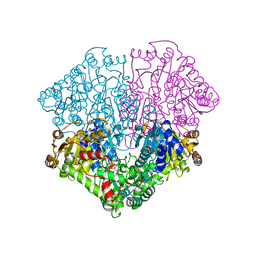 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-acetyl-thiamin diphosphate | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2E2E
 
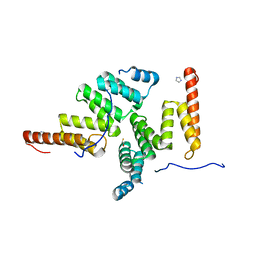 | | TPR domain of NrfG mediates the complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia Coli O157:H7 | | Descriptor: | BETA-MERCAPTOETHANOL, Formate-dependent nitrite reductase complex nrfG subunit, IMIDAZOLE | | Authors: | Han, D, Kim, K, Oh, J, Park, J, Kim, Y. | | Deposit date: | 2006-11-11 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TPR domain of NrfG mediates complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia coli O157:H7.
Proteins, 70, 2008
|
|
2E4E
 
 | |
2E4W
 
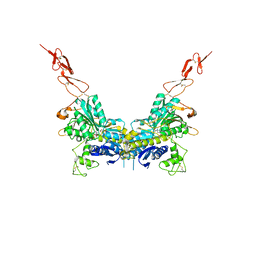 | | Crystal structure of the extracellular region of the group II metabotropic glutamate receptor complexed with 1S,3S-ACPD | | Descriptor: | (1S,3S)-1-aminocyclopentane-1,3-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EB5
 
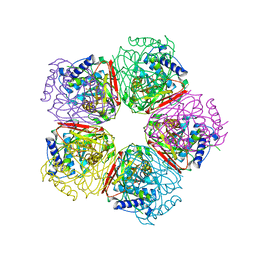 | | Crystal structure of HpcG complexed with oxalate | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2E4V
 
 | | Crystal structure of the extracellular region of the group II metabotropic glutamate receptor complexed with DCG-IV | | Descriptor: | (1R,2R)-3-[(S)-amino(carboxy)methyl]cyclopropane-1,2-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
