1ZYT
 
 | | Crystal structure of spin labeled T4 Lysozyme (A82R1) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | EPR (1.7 Å), X-RAY DIFFRACTION | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins
Protein Sci., 18, 2009
|
|
1XJO
 
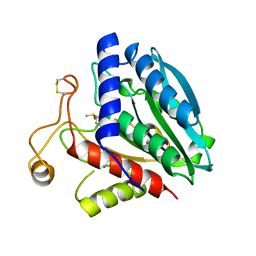 | | STRUCTURE OF AMINOPEPTIDASE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Greenblatt, H.M, Barra, D, Blumberg, S, Shoham, G. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Streptomyces griseus aminopeptidase: X-ray crystallographic structure at 1.75 A resolution.
J.Mol.Biol., 265, 1997
|
|
3TRA
 
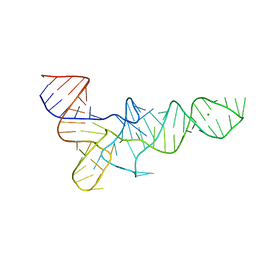 | |
2A7U
 
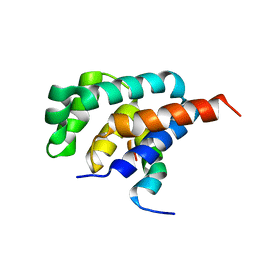 | | NMR solution structure of the E.coli F-ATPase delta subunit N-terminal domain in complex with alpha subunit N-terminal 22 residues | | Descriptor: | ATP synthase alpha chain, ATP synthase delta chain | | Authors: | Wilkens, S, Borchardt, D, Weber, J, Senior, A.E. | | Deposit date: | 2005-07-06 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Interaction of the delta and alpha Subunits of the Escherichia coli F(1)F(0)-ATP Synthase by NMR Spectroscopy
Biochemistry, 44, 2005
|
|
6TRV
 
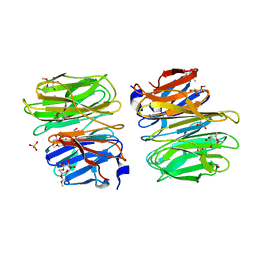 | |
6O31
 
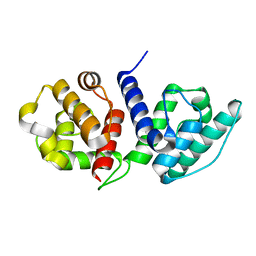 | |
6TPU
 
 | | Crystal structures of FNIII domain three and four of the human leucocyte common antigen-related protein, LAR | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4UXI
 
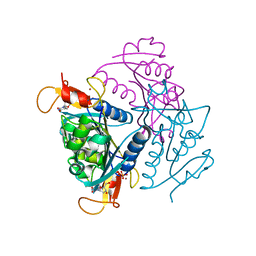 | | Leishmania major Thymidine Kinase in complex with thymidine | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
2YKG
 
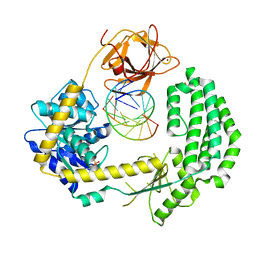 | | Structural insights into RNA recognition by RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
6NWA
 
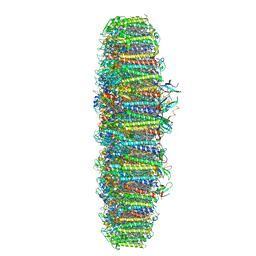 | | The structure of the photosystem I IsiA super-complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Li, J, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-02-06 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The structure of the stress-induced photosystem I-IsiA antenna supercomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1DAY
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF PROTEIN KINASE CK2 (ALPHA-SUBUNIT) AND MG-GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN KINASE CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.G, Schomburg, D. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GTP plus water mimic ATP in the active site of protein kinase CK2.
Nat.Struct.Biol., 6, 1999
|
|
2YU9
 
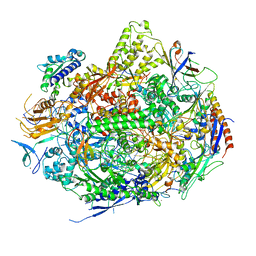 | | RNA polymerase II elongation complex in 150 mm MG+2 with UTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2007-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
8GAA
 
 | |
2Y8T
 
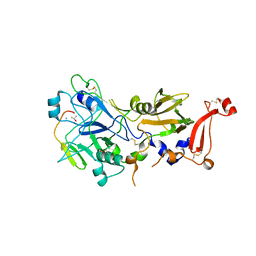 | | Co-structure of AMA1 with a surface exposed region of RON2 from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE, ... | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
2YW7
 
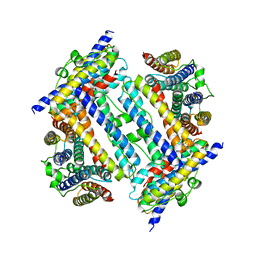 | | Crystal structure of C-terminal deletion mutant of Mycobacterium smegmatis Dps | | Descriptor: | Starvation-induced DNA protecting protein | | Authors: | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
2YDZ
 
 | | X-ray structure of the cyan fluorescent protein SCFP3A (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
4UUZ
 
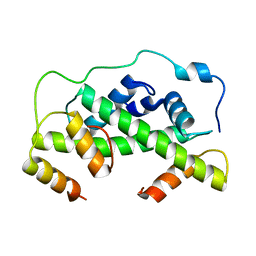 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
4V0Q
 
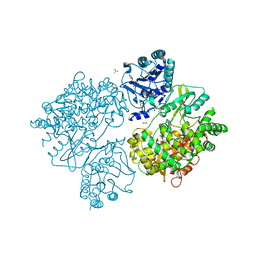 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
1DJE
 
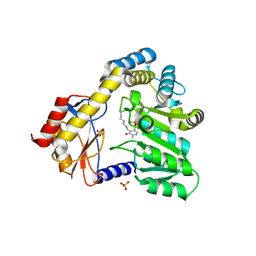 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF 8-AMINO-7-OXONANOATE SYNTHASE | | Descriptor: | 8-AMINO-7-OXONANOATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
4V2S
 
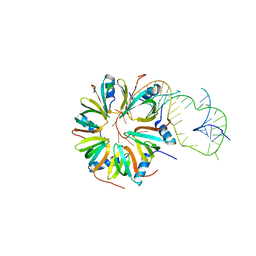 | | Crystal structure of Hfq in complex with the sRNA RydC | | Descriptor: | RNA-BINDING PROTEIN HFQ, RYDC | | Authors: | Dimastrogiovanni, D, Frohlich, K.S, Bruce, H.A, Bandyra, K.J, Hohensee, S, Vogel, J, Luisi, B.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Recognition of the small regulatory RNA RydC by the bacterial Hfq protein.
Elife, 3, 2014
|
|
4OYD
 
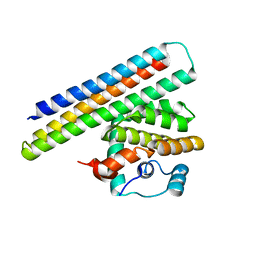 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
8GL3
 
 | |
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
6NYS
 
 | |
