2JWM
 
 | |
8CZV
 
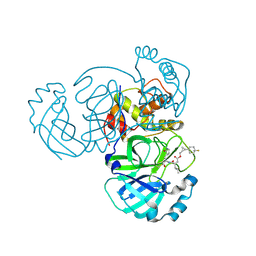 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
2FQC
 
 | | Solution structure of conotoxin pl14a | | Descriptor: | Alpha/kappa-conotoxin pl14a | | Authors: | Craik, D.J, Daly, N.L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-07-18 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A Novel Conotoxin Inhibitor of Kv1.6 Channel and nAChR Subtypes Defines a New Superfamily of Conotoxins
Biochemistry, 45, 2006
|
|
6VII
 
 | |
8P3B
 
 | | Neisseria meningitidis Type IV pilus SA-GATDH variant | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Fimbrial protein, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
5IIH
 
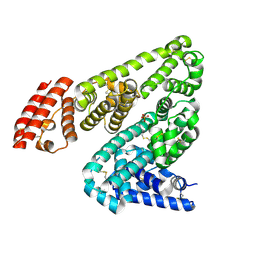 | | Crystal structure of Equine Serum Albumin in the presence of 2.5 mM zinc at pH 7.4 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
3EAR
 
 | |
7OI5
 
 | | Crystal structure of AP2 Mu2 - FCHO2 chimera (GST cleaved) | | Descriptor: | AP-2 complex subunit mu,F-BAR domain only protein 2, GLYCEROL | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
6VPM
 
 | |
6ZMP
 
 | |
1ALY
 
 | |
2J52
 
 | | Solution Structure of GB1 domain Protein G and low and high pressure. | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wilton, D.J, Tunnicliffe, R.B, Kamatari, Y.O, Akasaka, K, Williamson, M.P. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Proteins, 71, 2008
|
|
3EM0
 
 | | Crystal structure of Zebrafish Ileal Bile Acid-Bindin Protein complexed with cholic acid (crystal form B). | | Descriptor: | CHOLIC ACID, Ileal Bile Acid-Binding Protein | | Authors: | Capaldi, S, Saccomani, G, Fessas, D, Signorelli, M, Perduca, M, Monaco, H.L. | | Deposit date: | 2008-09-23 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-Ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule.
J.Mol.Biol., 385, 2009
|
|
3ETB
 
 | | Crystal structure of the engineered neutralizing antibody M18 complexed with anthrax protective antigen domain 4 | | Descriptor: | Anthrax Protective Antigen, Antibody M18 light chain and antibody M18 heavy chain linked with a synthetic (GGGGS)4 linker | | Authors: | Monzingo, A.F, Leysath, C.E, Barnett, J, Iverson, B.L, Georgiou, G, Robertus, J.D. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the engineered neutralizing antibody M18 complexed to domain 4 of the anthrax protective antigen.
J.Mol.Biol., 387, 2009
|
|
3EPE
 
 | |
1A4T
 
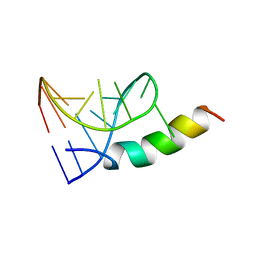 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
8T2B
 
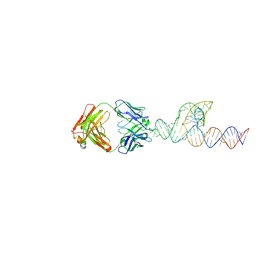 | |
8T2O
 
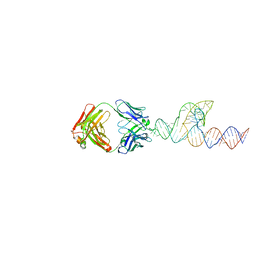 | |
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2025-04-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
8T2A
 
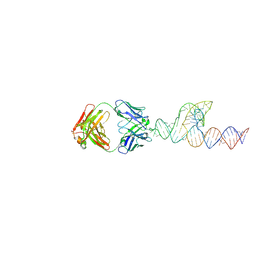 | |
2JUJ
 
 | |
8T29
 
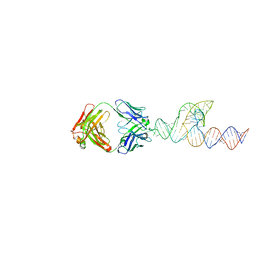 | | Crystal structure of SCV PTE RNA in complex with Fab BL3-6 | | Descriptor: | BL3-6 Fab heavy chain, BL3-6 Fab light chain, RNA (90-MER) | | Authors: | Ojha, M, Koirala, D. | | Deposit date: | 2023-06-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structure of saguaro cactus virus 3' translational enhancer mimics 5' cap for eIF4E binding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5W3N
 
 | | Molecular structure of FUS low sequence complexity domain protein fibrils | | Descriptor: | RNA-binding protein FUS | | Authors: | Murray, D.T, Kato, M, Lin, Y, Thurber, K, Hung, I, McKnight, S, Tycko, R. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains.
Cell, 171, 2017
|
|
7B7V
 
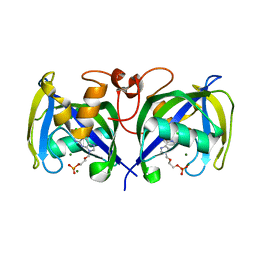 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
5VHS
 
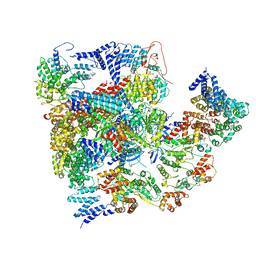 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
