1HH5
 
 | | cytochrome c7 from Desulfuromonas acetoxidans | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Czjzek, M, Haser, R, Arnoux, P, Shepard, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Cytochrome C7 from Desulfuromonas Acetoxidans at 1.9A Resolutio N
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AQE
 
 | |
5M41
 
 | | Crystal structure of nigritoxine | | Descriptor: | MAGNESIUM ION, Nigritoxine | | Authors: | Czjzek, M, Labreuche, L, Jeudy, A, Le Roux, F. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nigritoxin is a bacterial toxin for crustaceans and insects.
Nat Commun, 8, 2017
|
|
5OCQ
 
 | | Crystal structure of the complex of the kappa-carrageenase from Pseudoalteromonas carrageenovora with an oligotetrasaccharide of kappa-carrageenan | | Descriptor: | 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, CITRIC ACID, ... | | Authors: | Czjzek, M, Leroux, C, Bernard, T, Matard-Mann, M, Jeudy, A, Michel, G. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
2JFE
 
 | | The crystal structure of human cytosolic beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOSOLIC BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Tribolo, S, Berrin, J.G, Kroon, P.A, Juge, N. | | Deposit date: | 2007-01-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Human Cytosolic Beta-Glucosidase Unravels the Substrate Aglycone Specificity of a Family 1 Glycoside Hydrolase
J.Mol.Biol., 370, 2007
|
|
5OCR
 
 | | Crystal structure of the kappa-carrageenase zobellia_236 from Zobellia galactanivorans | | Descriptor: | GLYCEROL, Kappa-carrageenase, MAGNESIUM ION | | Authors: | Czjzek, M, Matard-Mann, M, Michel, G, Jeudy, A, Larocque, R. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into marine carbohydrate degradation by family GH16 kappa-carrageenases.
J. Biol. Chem., 292, 2017
|
|
1CZJ
 
 | | CYTOCHROME C OF CLASS III (AMBLER) 26 KD | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Czjzek, M, Haser, R. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a dimeric octaheme cytochrome c3 (M(r) 26,000) from Desulfovibrio desulfuricans Norway.
Structure, 4, 1996
|
|
2UYD
 
 | | Crystal structure of the SmHasA mutant H83A | | Descriptor: | ACETATE ION, HEMOPHORE HASA, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Czjzek, M, Caillet-Saguy, C, Fournelle, A, Guigliarelli, B, Izadi-Pruneyre, N, Lecroisey, A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deciphering the Structural Role of Histidine 83 for Heme Binding in Hemophore Hasa.
J.Biol.Chem., 283, 2008
|
|
2BS9
 
 | | Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus | | Descriptor: | BETA-XYLOSIDASE, CALCIUM ION | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
2BFG
 
 | | crystal structure of beta-xylosidase (fam GH39) in complex with dinitrophenyl-beta-xyloside and covalently bound xyloside | | Descriptor: | 2,5-DINITROPHENOL, BETA-XYLOSIDASE, SODIUM ION, ... | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
2CY3
 
 | |
2CN4
 
 | | The crystal structure of the secreted dimeric form of the hemophore HasA reveals a domain swapping with an exchanged heme ligand | | Descriptor: | HEMOPHORE HASA, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Czjzek, M, Letoffe, S, Wandersman, C, Delepierre, M, Lecroisey, A, Izadi-Pruneyre, N. | | Deposit date: | 2006-05-18 | | Release date: | 2006-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Secreted Dimeric Form of the Hemophore Hasa Reveals a Domain Swapping with an Exchanged Heme Ligand
J.Mol.Biol., 365, 2007
|
|
7BJT
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | Alginate lyase, family PL17, CALCIUM ION, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
7BM6
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid, Alginate lyase, family PL17, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
1UY0
 
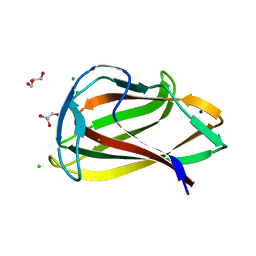 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYX
 
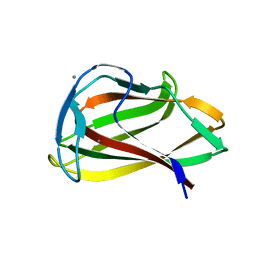 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYZ
 
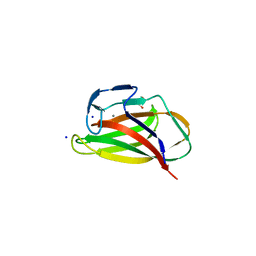 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UXX
 
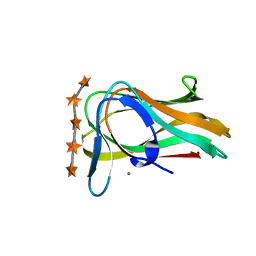 | | CBM6ct from Clostridium thermocellum in complex with xylopentaose | | Descriptor: | CALCIUM ION, XYLANASE U, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Henrissat, D.B.B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UXZ
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
2VH9
 
 | | CRYSTAL STRUCTURE OF NXG1-DELTAYNIIG IN COMPLEX WITH XLLG, A XYLOGLUCAN DERIVED OLIGOSACCHARIDE | | Descriptor: | CELLULASE, GLYCEROL, ZINC ION, ... | | Authors: | Czjzek, M, Mark, P, Baumann, M.J, Eklof, J.M, Michel, G, Brumer, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of Nasturtium Tmnxg1 Complexes by Crystallography and Molecular Dynamics Provides Detailed Insight Into Substrate Recognition by Family Gh16 Xyloglucan Endo-Transglycosylases and Endo-Hydrolases.
Proteins, 75, 2009
|
|
1H49
 
 | | CRYSTAL STRUCTURE OF THE INACTIVE DOUBLE MUTANT OF THE MAIZE BETA-GLUCOSIDASE ZMGLU1-E191D-F198V IN COMPLEX WITH DIMBOA-GLUCOSIDE | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Moriniere, J, Verdoucq, L, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and Structural Analysis of Aglycone Specificity in Maize and Sorghum Beta-Glucosidases
J.Biol.Chem., 278, 2003
|
|
2BQ4
 
 | | Crystal structure of type I cytochrome c3 from Desulfovibrio africanus | | Descriptor: | BASIC CYTOCHROME C3, CALCIUM ION, HEME C | | Authors: | Czjzek, M, Pieulle, L, Morelli, X, Guerlesquin, F, Hatchikian, E.C. | | Deposit date: | 2005-04-27 | | Release date: | 2005-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Type I / Type II Cytochrome C(3) Complex: An Electron Transfer Link in the Hydrogen-Sulfate Reduction Pathway.
J.Mol.Biol., 354, 2005
|
|
1EGZ
 
 | | CELLULASE CEL5 FROM ERWINIA CHRYSANTHEMI, A FAMILY GH 5-2 ENZYME | | Descriptor: | CALCIUM ION, ENDOGLUCANASE Z | | Authors: | Czjzek, M, El Hassouni, M, Py, B, Barras, F. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II protein secretion in gram-negative pathogenic bacteria: the study of the structure/secretion relationships of the cellulase Cel5 (formerly EGZ) from Erwinia chrysanthemi
J.Mol.Biol., 310, 2001
|
|
