3CIC
 
 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CID
 
 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4DJW
 
 | |
4DJU
 
 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJX
 
 | |
4DJY
 
 | |
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPK
 
 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LNK
 
 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPJ
 
 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NEH
 
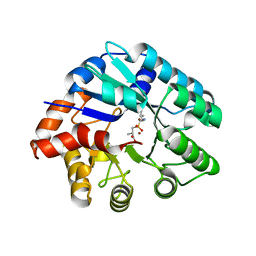 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3S2N
 
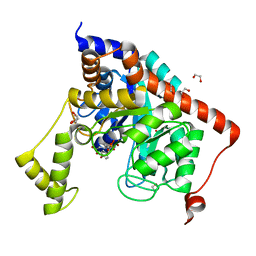 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Tyr-D-Asp | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-2-(4-hydroxyphenyl)ethyl](hydroxy)phosphoryl]methyl}butanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Tyr-D-Asp
To be Published
|
|
3S2M
 
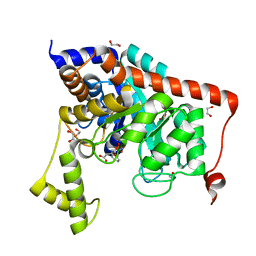 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Phe-D-Asp | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]methyl}butanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Phe-D-Asp
To be Published
|
|
3GIP
 
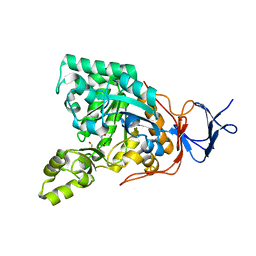 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3GIQ
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3ID7
 
 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3LY0
 
 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
3S2L
 
 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Glu | | Descriptor: | (2R)-2-{[(S)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Glu
To be Published
|
|
3S2J
 
 | | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Crystal structure of dipeptidase from Streptomyces coelicolor complexed with phosphinate pseudodipeptide L-Leu-D-Ala
To be Published
|
|
3K5X
 
 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
