7AD2
 
 | |
7A3W
 
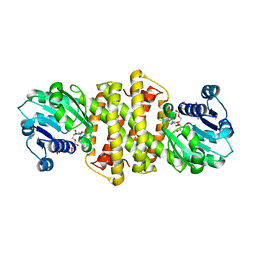 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
5FR9
 
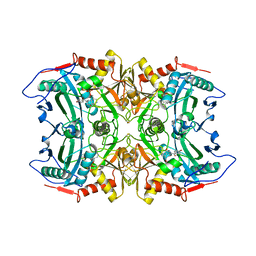 | | Structure of transaminase ATA-117 arRmut11 from Arthrobacter sp. KNK168 inhibited with 1-(4-Bromophenyl)-2-fluoroethylamine | | Descriptor: | (R)-AMINE TRANSAMINASE, [4-[3-(4-bromophenyl)-3-oxidanylidene-propyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl phosphate | | Authors: | Cuetos, A, Kroutil, W, Lavandera, I, Grogan, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Catalytic Promiscuity of Transaminases: Preparation of Enantioenriched Beta-Fluoroamines by Formal Tandem Hydrodefluorination/Deamination.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5G4J
 
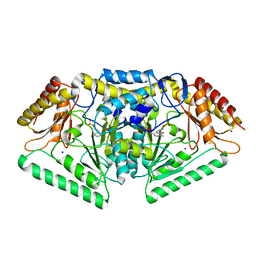 | | Phospholyase A1RDF1 from Arthrobacter in complex with phosphoethanolamine | | Descriptor: | PUTATIVE AMINOTRANSFERASE CLASS III PROTEIN, SODIUM ION, {5-hydroxy-6-methyl-4-[(E)-{[2-(phosphonooxy)ethyl]imino}methyl]pyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
5G4I
 
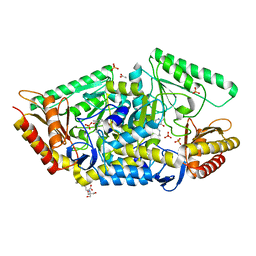 | | PLP-dependent phospholyase A1RDF1 from Arthrobacter aurescens TC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
6T9H
 
 | |
6TFN
 
 | |
6TFT
 
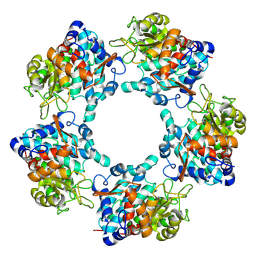 | |
6THM
 
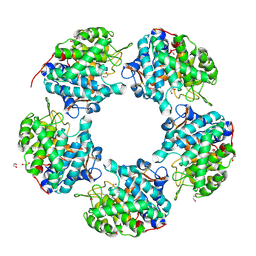 | | Linalool Dehydratase Isomerase M125A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI, MALONATE ION | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6TFR
 
 | | Linalool Dehydratase Isomerase C180A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6H1B
 
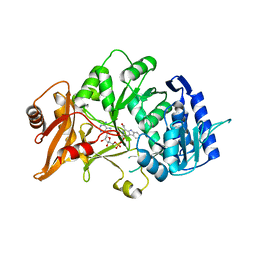 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6RZ2
 
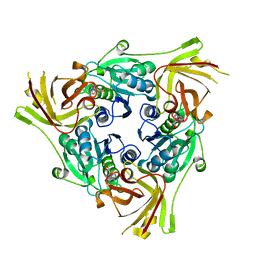 | | SalL with Chloroadenosine | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Adenosyl-chloride synthase | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6RYZ
 
 | | SalL with S-adenosyl methionine | | Descriptor: | 1,2-ETHANEDIOL, Adenosyl-chloride synthase, CHLORIDE ION, ... | | Authors: | McKean, I, Frese, A, Cuetos, A, Burley, G, Grogan, G. | | Deposit date: | 2019-06-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
