2NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND WATER BOUND IN ACTIVE CENTER | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
2NOS
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114), AMINOGUANIDINE COMPLEX | | Descriptor: | AMINOGUANIDINE, IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
2HP7
 
 | | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor | | Descriptor: | Flagellar motor switch protein FliM | | Authors: | Crane, B.R, Park, S, Lowder, B, Bilwes, A.M, Blair, D.F. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7UD0
 
 | | Room Temperature Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Room-temperature serial synchrotron crystallography of Drosophila cryptochrome.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6R9N
 
 | | Peroxy diiron species of chemotaxis sensor ODP | | Descriptor: | CHLORIDE ION, FE (III) ION, OXYGEN ATOM, ... | | Authors: | Muok, A.R, Crane, B.R. | | Deposit date: | 2019-04-03 | | Release date: | 2019-06-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4QYW
 
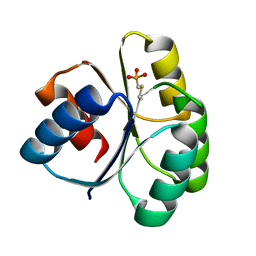 | |
1R1C
 
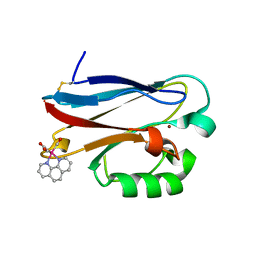 | | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (I) ION | | Authors: | Miller, J.E, Gradinaru, C, Crane, B.R, Di Bilio, A.J. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment
J.Am.Chem.Soc., 125, 2003
|
|
6WTB
 
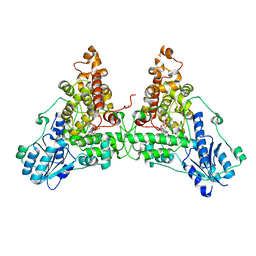 | | Sort-Tagged Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Tuning flavin environment to detect and control light-induced conformational switching in Drosophila cryptochrome.
Commun Biol, 4, 2021
|
|
6Y1Y
 
 | |
4JPB
 
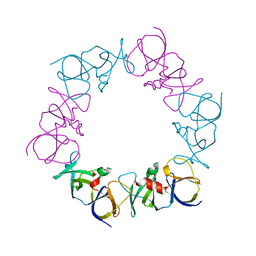 | | The structure of a ternary complex between CheA domains P4 and P5 with CheW and with an unzipped fragment of TM14, a chemoreceptor analog from Thermotoga maritima. | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein | | Authors: | Li, X, Bayas, C, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2013-03-19 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | The 3.2 angstrom resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays.
Biochemistry, 52, 2013
|
|
5TDY
 
 | | Structure of cofolded FliFc:FliGn complex from Thermotoga maritima | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG | | Authors: | Lynch, M.J, Levenson, R, Kim, E.A, Sircar, R, Blair, D.F, Dahlquist, F.W, Crane, B.R. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Structure, 25, 2017
|
|
3T8Y
 
 | |
2B12
 
 | |
2AN2
 
 | | P332G, A333S Double mutant of the Bacillus subtilis Nitric Oxide Synthase | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, P332G A333S double mutant of Nitric Oxide Synthase from Bacillus subtilis, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a loose dimer: an intermediate in nitric oxide synthase assembly
J.Mol.Biol., 352, 2005
|
|
3SOH
 
 | | Architecture of the Flagellar Rotor | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Koushik, P, Gonzalez-Bonet, G, Bilwes, A.M, Crane, B.R, Blair, D. | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the flagellar rotor.
Embo J., 30, 2011
|
|
4XIV
 
 | |
3UR1
 
 | |
9C8O
 
 | |
9C8M
 
 | |
9C8L
 
 | |
9C8P
 
 | |
3LNR
 
 | |
8DIK
 
 | |
8DD7
 
 | | The Cryo-EM structure of Drosophila Cryptochrome in complex with Timeless | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylated-DNA--protein-cysteine methyltransferase,Cryptochrome-1 fusion, Protein timeless,Methylated-DNA--protein-cysteine methyltransferase fusion | | Authors: | Feng, S, Lin, C, DeOliveira, C.C, Crane, B.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Nature, 617, 2023
|
|
6C40
 
 | | CheY41PyTyrD54K from Thermotoga maritima | | Descriptor: | COPPER (II) ION, Chemotaxis protein CheY | | Authors: | Merz, G.E, Muok, A.R, Crane, B.R. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Site-Specific Incorporation of a Cu2+Spin Label into Proteins for Measuring Distances by Pulsed Dipolar Electron Spin Resonance Spectroscopy.
J Phys Chem B, 122, 2018
|
|
