1RO2
 
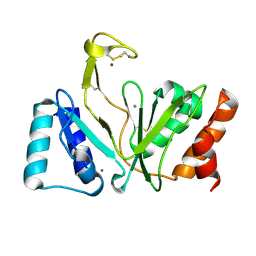 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M manganese soak | | 分子名称: | MANGANESE (II) ION, ZINC ION, hypothetical protein ORF904 | | 著者 | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | 登録日 | 2003-12-01 | | 公開日 | 2004-01-27 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T9Z
 
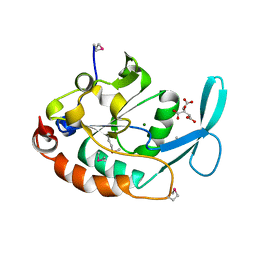 | | Three-dimensional structure of a RNA-polymerase II binding protein. | | 分子名称: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | 著者 | Kamenski, T, Heilmeier, S, Meinhart, A, Cramer, P. | | 登録日 | 2004-05-19 | | 公開日 | 2004-08-31 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Mechanism of
RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
1SZ9
 
 | |
1TA0
 
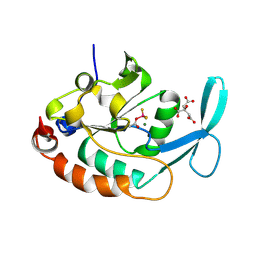 | | Three-dimensional structure of a RNA-polymerase II binding protein with associated ligand. | | 分子名称: | CITRIC ACID, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | 著者 | Kamenski, T, Heilmeier, S, Meinhart, T, Cramer, P. | | 登録日 | 2004-05-19 | | 公開日 | 2004-08-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and Mechanism of RNA Polymerase II CTD Phosphatases.
Mol.Cell, 15, 2004
|
|
6RFL
 
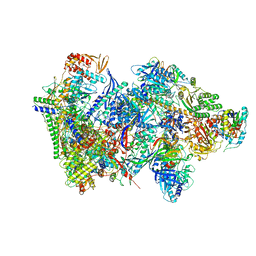 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | 分子名称: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | 著者 | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | 登録日 | 2019-04-15 | | 公開日 | 2019-12-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
3QWD
 
 | | Crystal structure of ClpP from Staphylococcus aureus | | 分子名称: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | 著者 | Geiger, S.R, Boettcher, T, Sieber, S.A, Cramer, P. | | 登録日 | 2011-02-28 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Conformational Switch Underlies ClpP Protease Function.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6T9L
 
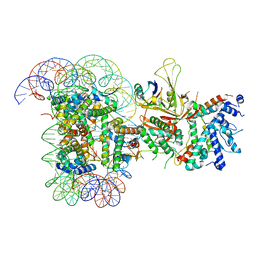 | |
6TDA
 
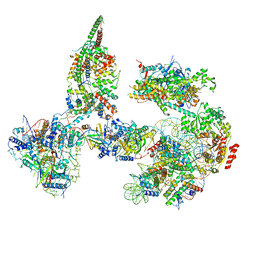 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | 著者 | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | 登録日 | 2019-11-08 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
1YKE
 
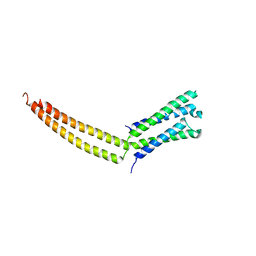 | |
6RYU
 
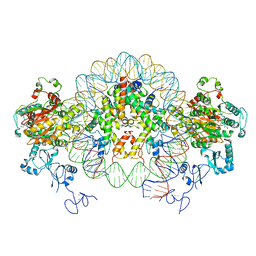 | | Nucleosome-CHD4 complex structure (two CHD4 copies) | | 分子名称: | Chromodomain-helicase-DNA-binding protein 4,CHD4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | 著者 | Farnung, L, Ochmann, M, Cramer, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
3QT1
 
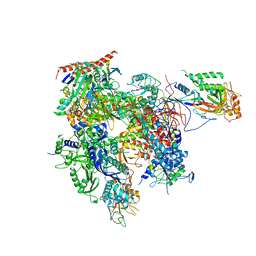 | | RNA polymerase II variant containing A Chimeric RPB9-C11 subunit | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Ruan, W, Lehmann, E, Thomm, M, Kostrewa, D, Cramer, P. | | 登録日 | 2011-02-22 | | 公開日 | 2011-03-23 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Evolution of two modes of intrinsic RNA polymerase transcript cleavage.
J.Biol.Chem., 286, 2011
|
|
6RO4
 
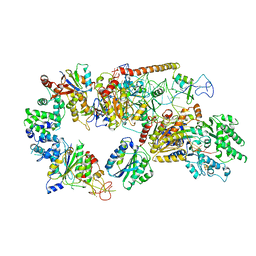 | | Structure of the core TFIIH-XPA-DNA complex | | 分子名称: | DNA repair protein complementing XP-A cells, DNA1, DNA2, ... | | 著者 | Kokic, G, Chernev, A, Tegunov, D, Dienemann, C, Urlaub, H, Cramer, P. | | 登録日 | 2019-05-10 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of TFIIH activation for nucleotide excision repair.
Nat Commun, 10, 2019
|
|
3M1M
 
 | | Crystal structure of the primase-polymerase from Sulfolobus islandicus | | 分子名称: | GLYCEROL, ORF904, SULFATE ION, ... | | 著者 | Vannini, A, Beck, K, Lipps, G, Cramer, P. | | 登録日 | 2010-03-05 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis
Nucleic Acids Res., 38, 2010
|
|
7PET
 
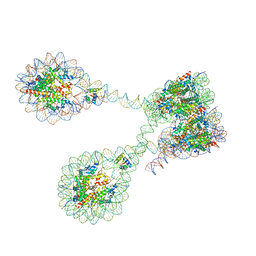 | |
7O75
 
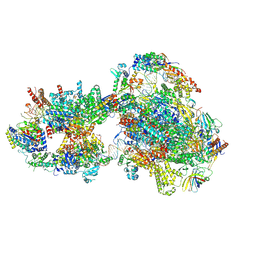 | | Yeast RNA polymerase II transcription pre-initiation complex with open promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-13 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4L
 
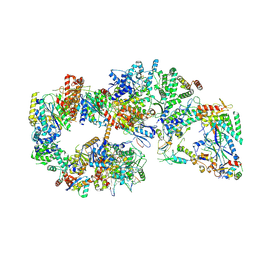 | | Yeast TFIIH in the expanded state within the pre-initiation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-06 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4K
 
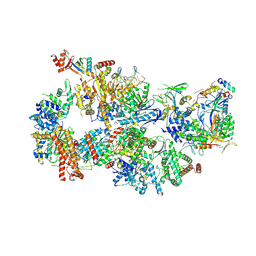 | | Yeast TFIIH in the contracted state within the pre-initiation complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-06 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O72
 
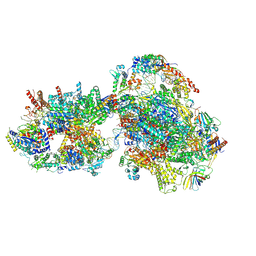 | | Yeast RNA polymerase II transcription pre-initiation complex with closed promoter DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | 登録日 | 2021-04-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
3GXX
 
 | | Structure of the SH2 domain of the Candida glabrata transcription elongation factor Spt6, crystal form B | | 分子名称: | Transcription elongation factor SPT6 | | 著者 | Dengl, S, Mayer, A, Sun, M, Cramer, P. | | 登録日 | 2009-04-03 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and in vivo requirement of the yeast Spt6 SH2 domain
J.Mol.Biol., 389, 2009
|
|
6RYR
 
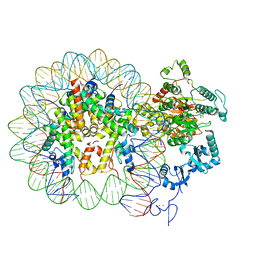 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | 分子名称: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | 著者 | Farnung, L, Ochmann, M, Cramer, P. | | 登録日 | 2019-06-11 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
3GXW
 
 | | Structure of the SH2 domain of the Candida glabrata transcription elongation factor Spt6, crystal form A | | 分子名称: | SODIUM ION, SUCCINIC ACID, Transcription elongation factor SPT6 | | 著者 | Dengl, S, Mayer, A, Sun, M, Cramer, P. | | 登録日 | 2009-04-03 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and in vivo requirement of the yeast Spt6 SH2 domain
J.Mol.Biol., 389, 2009
|
|
6S01
 
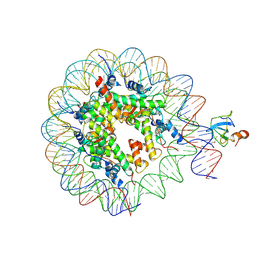 | | Structure of LEDGF PWWP domain bound H3K36 methylated nucleosome | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | 著者 | Wang, H, Farnung, L, Dienemann, C, Cramer, P. | | 登録日 | 2019-06-13 | | 公開日 | 2019-12-18 | | 最終更新日 | 2020-01-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of H3K36-methylated nucleosome-PWWP complex reveals multivalent cross-gyre binding.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7PEU
 
 | |
7PF2
 
 | |
7PFT
 
 | |
