6F7E
 
 | |
6TWE
 
 | |
5LW4
 
 | |
6Z41
 
 | |
6Z40
 
 | |
6XTH
 
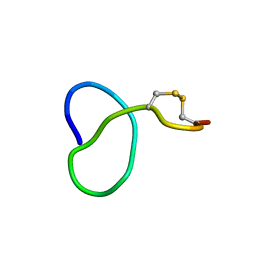 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A1 | | Authors: | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTI
 
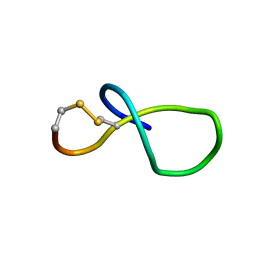 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A2 | | Authors: | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
