4LDV
 
 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDX
 
 | | Crystal structure of the DNA binding domain of arabidopsis thaliana auxin response factor 1 (ARF1) in complex with protomor-like sequence ER7 | | Descriptor: | Auxin response factor 1, ER7, forward sequence, ... | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
4D7A
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with AMP at 1.801 Angstroem resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
4D79
 
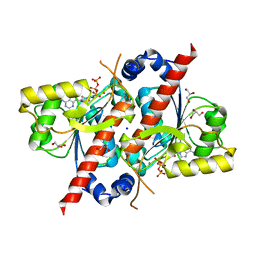 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
1UDR
 
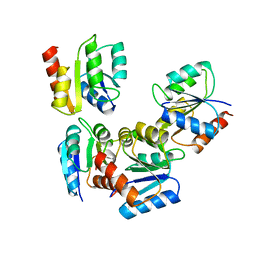 | |
2RNF
 
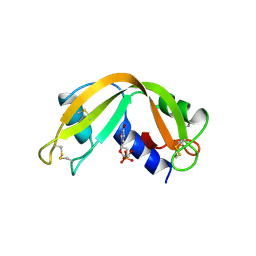 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
1OMH
 
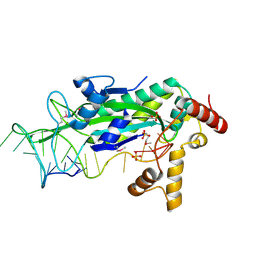 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
1OSB
 
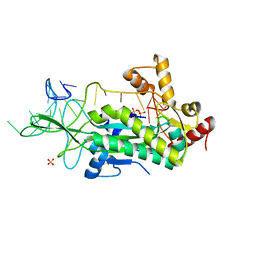 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | Dna oligonucleotide, SULFATE ION, TrwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
4CVQ
 
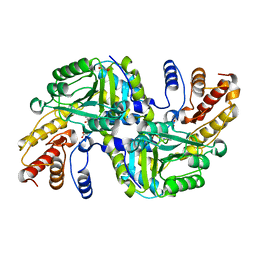 | | CRYSTAL STRUCTURE OF AN AMINOTRANSFERASE FROM ESCHERICHIA COLI AT 2. 11 ANGSTROEM RESOLUTION | | Descriptor: | ACETATE ION, GLUTAMATE-PYRUVATE AMINOTRANSFERASE ALAA, GLYCEROL, ... | | Authors: | Penya-Soler, E, Fernandez, F.J, Lopez-Estepa, M, Garces, F, Richardson, A.J, Rudd, K.E, Coll, M, Vega, M.C. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural analysis and mutant growth properties reveal distinctive enzymatic and cellular roles for the three major L-alanine transaminases of Escherichia coli.
PLoS ONE, 9, 2014
|
|
467D
 
 | |
1NDN
 
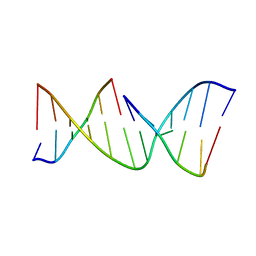 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
2JB9
 
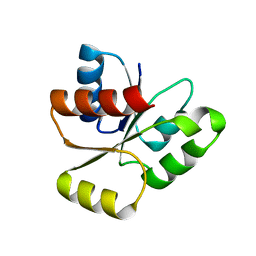 | | PhoB response regulator receiver domain constitutively-active double mutant D10A and D53E. | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Ferrer-Orta, C, Arribas-Bosacoma, R, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the E. Coli Phob Receiver Domain Give Insights Into Activation
J.Mol.Biol., 366, 2007
|
|
2JBA
 
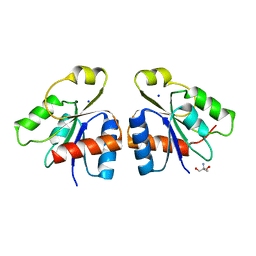 | | PhoB response regulator receiver domain constitutively-active double mutant D53A and Y102C. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB, SODIUM ION | | Authors: | Arribas-Bosacoma, R, Ferrer-Orta, C, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the Escherichia Coli Phob Receiver Domain Give Insights Into Activation.
J.Mol.Biol., 366, 2007
|
|
1YMU
 
 | |
1YMV
 
 | |
1VTE
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
2CPG
 
 | | TRANSCRIPTIONAL REPRESSOR COPG | | Descriptor: | CHLORIDE ION, TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Sola, M, Acebo, P, Parraga, A, Guasch, A, Eritja, R, Gonzalez, A, Espinosa, M, del Solar, G, Coll, M. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator.
EMBO J., 17, 1998
|
|
264D
 
 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
2CDM
 
 | | The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*CP*CP*GP*AP*AP *AP*GP*GP*TP*GP*CP*GP*TP*AP*TP*TP*GP*TP*CP*TP*AP*T)-3', SULFATE ION, TRWC | | Authors: | Boer, R, Russi, S, Guasch, A, Lucas, M, Blanco, A.G, Perez-Luque, R, Coll, M, de la Cruz, F. | | Deposit date: | 2006-01-25 | | Release date: | 2006-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unveiling the Molecular Mechanism of a Conjugative Relaxase: The Structure of Trwc Complexed with a 27-mer DNA Comprising the Recognition Hairpin and the Cleavage Site.
J.Mol.Biol., 358, 2006
|
|
4A00
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
1K9G
 
 | | Crystal Structure of the Complex of Cryptolepine-d(CCTAGG)2 | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*G)-3', 5-METHYL-5H-INDOLO[3,2-B]QUINOLINE | | Authors: | Lisgarten, J.N, Coll, M, Portugal, J, Wright, C.W, Aymami, J. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The antimalarial and cytotoxic drug cryptolepine intercalates into DNA at cytosine-cytosine sites.
Nat.Struct.Biol., 9, 2002
|
|
1KWM
 
 | | Human procarboxypeptidase B: Three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI) | | Descriptor: | CITRIC ACID, Procarboxypeptidase B, ZINC ION | | Authors: | Pereira, P.J.B, Segura-Martin, S, Ferrer-Orta, C, Vendrell, J, Aviles, F.-X, Coll, M, Gomis-Rueth, F.-X. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human procarboxypeptidase B: three-dimensional structure and implications for thrombin-activatable fibrinolysis inhibitor (TAFI).
J.Mol.Biol., 321, 2002
|
|
