3PXN
 
 | | Crystal structure of the Drosophila kinesin family member Kin10/NOD in complex with divalent manganese and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein Nod, MANGANESE (II) ION | | Authors: | Cochran, J.C, Zhao, Y.C, Wilcox, D.E, Kull, F.J. | | Deposit date: | 2010-12-10 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A metal switch for controlling the activity of molecular motor proteins.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3DC4
 
 | | Crystal structure of the Drosophila kinesin family member NOD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein Nod, MAGNESIUM ION | | Authors: | Cochran, J.C, Mulko, N.K, Kull, F.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement.
Cell(Cambridge,Mass.), 136, 2009
|
|
3DCB
 
 | | Crystal structure of the Drosophila kinesin family member NOD in complex with AMPPNP | | Descriptor: | Kinesin-like protein Nod, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cochran, J.C, Mulko, N.K, Kull, F.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement.
Cell(Cambridge,Mass.), 136, 2009
|
|
2NZ4
 
 | |
3J8Y
 
 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
3J8X
 
 | | High-resolution structure of no-nucleotide kinesin on microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
3DCO
 
 | | Drosophila NOD (3DC4) and Bovine Tubulin (1JFF) Docked into the 11-Angstrom Cryo-EM Map of Nucleotide-Free NOD Complexed to the Microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bovine Alpha Tubulin, Bovine Beta Tubulin, ... | | Authors: | Sindelar, C.V, Cochran, J.C, Kull, F.J. | | Deposit date: | 2008-06-04 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement.
Cell(Cambridge,Mass.), 136, 2009
|
|
6NJE
 
 | | Crystal structure of the motor domain of human kinesin family member 22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF22, ... | | Authors: | Walker, B.C, Zhu, H, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Park, H, Cochran, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the motor domain of human kinesin family member 22
To Be Published
|
|
1U9P
 
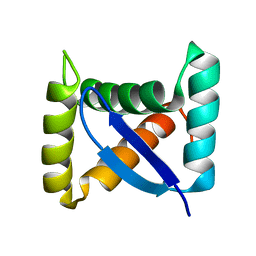 | | Permuted single-chain Arc | | Descriptor: | pArc | | Authors: | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3L3C
 
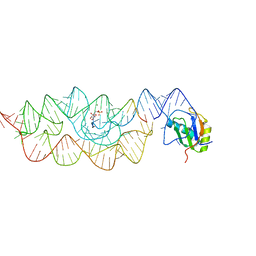 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to Glc6P | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G9C
 
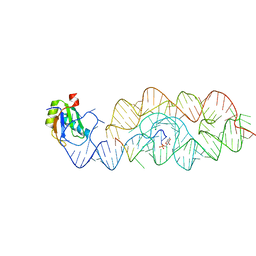 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G8T
 
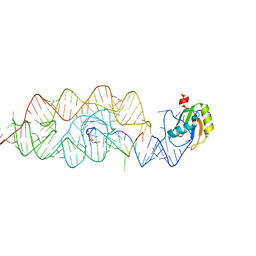 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G96
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to MaN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G8S
 
 | | Crystal structure of the pre-cleaved Bacillus anthracis glmS ribozyme | | Descriptor: | GLMS RIBOZYME, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3IIN
 
 | | Plasticity of the kink turn structural motif | | Descriptor: | DNA/RNA (5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*GP*AP*CP*C)-D(P*AP*GP*A)-R(P*CP*GP*GP*CP*C)-3'), DNA/RNA (5'-R(*CP*A)-D(P*T)-3'), Group I intron, ... | | Authors: | Lipchock, S.V, Strobel, S.A, Antonioli, A.H, Cochrane, J.C. | | Deposit date: | 2009-08-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Plasticity of the RNA kink turn structural motif.
Rna, 16, 2010
|
|
