1AD6
 
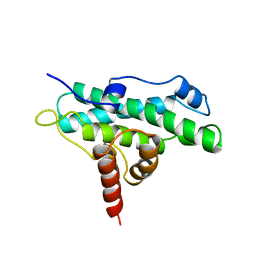 | | DOMAIN A OF HUMAN RETINOBLASTOMA TUMOR SUPPRESSOR | | Descriptor: | RETINOBLASTOMA TUMOR SUPPRESSOR | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 1997-02-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box.
Nat.Struct.Biol., 4, 1997
|
|
1A77
 
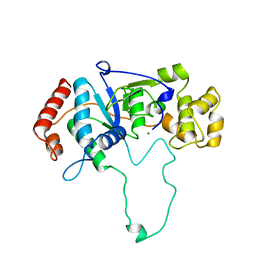 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1B78
 
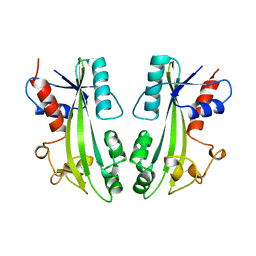 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
1B8V
 
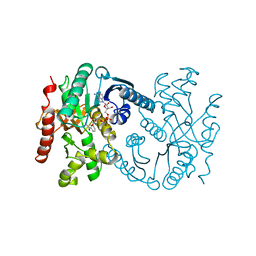 | | Malate dehydrogenase from Aquaspirillum arcticum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8U
 
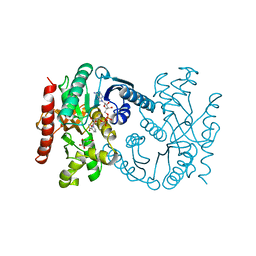 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8P
 
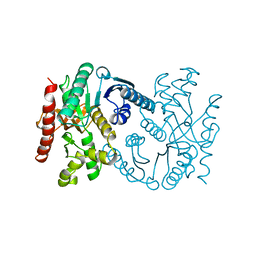 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1A76
 
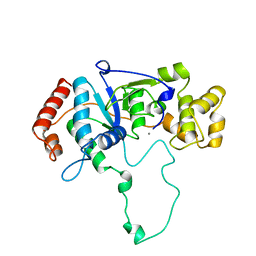 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
2ZIX
 
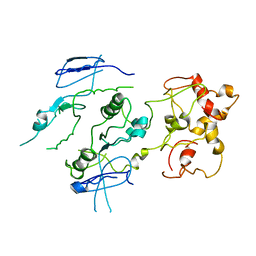 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIW
 
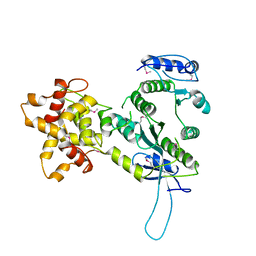 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
3A1J
 
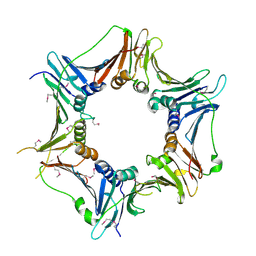 | | Crystal structure of the human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Sohn, S.Y, Cho, Y. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp
J.Mol.Biol., 390, 2009
|
|
2ZIU
 
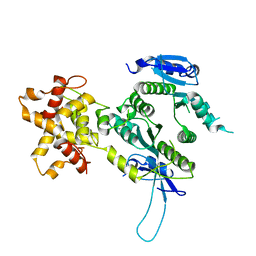 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
3AV0
 
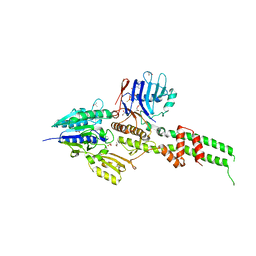 | | Crystal structure of Mre11-Rad50 bound to ATP S | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, GLYCEROL, ... | | Authors: | Lim, H.S, Kim, J.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
2ZIV
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
3AUZ
 
 | | Crystal structure of Mre11 with manganese | | Descriptor: | DNA double-strand break repair protein mre11, GLYCEROL, MANGANESE (II) ION | | Authors: | Park, Y.B, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUY
 
 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUX
 
 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex:Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
