3WCS
 
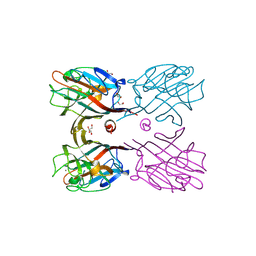 | | Crystal structure of plant lectin (ligand-bound form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
1UJJ
 
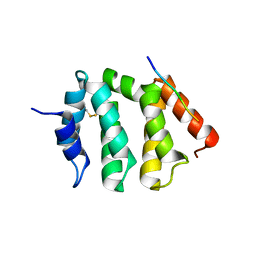 | | VHS domain of human GGA1 complexed with C-terminal peptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
1UJK
 
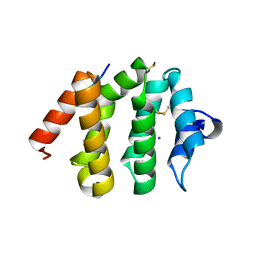 | | VHS domain of human GGA1 complexed with C-terminal phosphopeptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase, IODIDE ION | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
1WUH
 
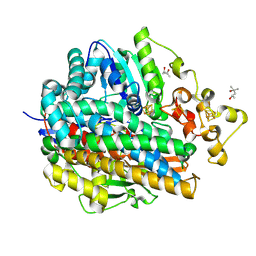 | | Three-Dimensional Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis for the activation process of [NiFe] hydrogenase from D.vulgaris Miyazaki F
To be Published
|
|
1WUJ
 
 | | Three-Dimensional Structure Of The Ni-B State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WUK
 
 | | High resolution Structure Of The Oxidized State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WXS
 
 | | Solution Structure of Ufm1, a ubiquitin-fold modifier | | Descriptor: | Ubiquitin-fold Modifier 1 | | Authors: | Sasakawa, H, Sakata, E, Yamaguchi, Y, Komatsu, M, Tatsumi, K, Kominami, E, Tanaka, K, Kato, K. | | Deposit date: | 2005-02-01 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1
Biochem.Biophys.Res.Commun., 343, 2006
|
|
1WUL
 
 | | High Resolution Structure Of The Reduced State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
1WYG
 
 | | Crystal Structure of a Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Hori, H, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: IDENTIFICATION OF THE TWO CYSTEINE DISULFIDE BONDS AND CRYSTAL STRUCTURE OF A NON-CONVERTIBLE RAT LIVER XANTHINE DEHYDROGENASE MUTANT
J.Biol.Chem., 280, 2005
|
|
1WUI
 
 | | Ultra-High resolution Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Activation process of [NiFe] hydrogenase elucidated by high-resolution X-Ray analyses: conversion of the ready to the unready state
Structure, 13, 2005
|
|
7CKE
 
 | |
7CKD
 
 | |
7DGN
 
 | | The Co-bound dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | COBALT (II) ION, Myoglobin, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGJ
 
 | | The dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGM
 
 | | The dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGL
 
 | | The Ni-bound dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | Myoglobin, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGO
 
 | | The Zn-bound dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGK
 
 | | The Co-bound dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | COBALT (II) ION, Myoglobin, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
6K7C
 
 | | Dimeric Shewanella violacea cytochrome c5 | | Descriptor: | HEME C, NITRATE ION, Soluble cytochrome cA | | Authors: | Yang, H, Yamanaka, M, Nagao, S, Yasuhara, K, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
7ECK
 
 | | RNA duplex containing C-Ag-A and U-Ag-A base pairs | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | RNA duplex containing C-Ag-A and U-Ag-A base pairs
To Be Published
|
|
7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
7ECL
 
 | | RNA duplex containing C-Ag-A and U-Ag-A base pair | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | RNA duplex containing C-Ag-A and U-Ag-A base pair
To Be Published
|
|
7ECJ
 
 | | RNA duplex containing C-A base pairs | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3') | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RNA duplex containing C-A base pairs
To Be Published
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
2ZMA
 
 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
