4BA5
 
 | | Crystal structure of omega-transaminase from Chromobacterium violaceum | | 分子名称: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, SULFATE ION | | 著者 | Sayer, C, Isupov, M.N, Littlechild, J.A. | | 登録日 | 2012-09-11 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Studies with Pseudomonas and Chromobacterium [Omega]-Aminotransferases Provide Insights Into Their Differing Substrate Specificity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4CF4
 
 | |
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | 分子名称: | L-HALOACID DEHALOGENASE, SULFATE ION | | 著者 | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | 登録日 | 2012-10-09 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | 分子名称: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | 著者 | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | 登録日 | 2008-02-29 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2ZLF
 
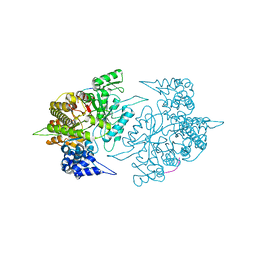 | | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase | | 分子名称: | FTLDADF, Ribonucleoside-diphosphate reductase large chain 1 | | 著者 | Xu, H, Fairman, J.W, Wijerathna, S.R, LaMacchia, J, Kreischer, N.R, Helmbrecht, E, Cooperman, B.S, Dealwis, C. | | 登録日 | 2008-04-09 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase: A Conformationally Flexible Pharmacophore
J.Med.Chem., 51, 2008
|
|
3ANO
 
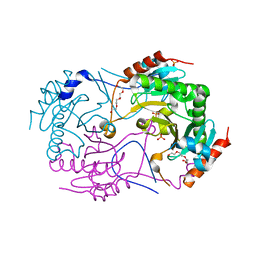 | | Crystal Structure of a Novel Diadenosine 5',5'''-P1,P4-Tetraphosphate Phosphorylase from Mycobacterium tuberculosis H37Rv | | 分子名称: | AP-4-A phosphorylase, PHOSPHATE ION, TETRAETHYLENE GLYCOL | | 著者 | Mori, S, Shibayama, K, Wachino, J, Arakawa, Y. | | 登録日 | 2010-09-06 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.894 Å) | | 主引用文献 | Structural insights into the novel diadenosine 5',5-P1,P4-tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv
J.Mol.Biol., 410, 2011
|
|
2ZLG
 
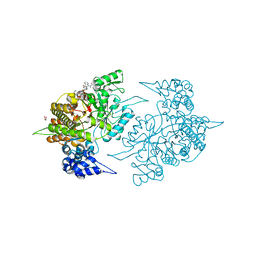 | | The Structual Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase | | 分子名称: | (5R,9S,12S,15S,18S,21S)-21-benzyl-12,18-bis(carboxymethyl)-15-cyclohexyl-1-(9H-fluoren-9-yl)-4-methyl-9-(2-methylpropyl)-3,6,10,13,16,19-hexaoxo-5-phenyl-2-oxa-4,8,11,14,17,20-hexaazadocosan-22-oic acid, GLYCEROL, Ribonucleoside-diphosphate reductase large chain 1 | | 著者 | Xu, H, Fairman, J.W, Wijerathna, S.R, LaMacchia, J, Kreischer, N.R, Helmbrecht, E, Cooperman, B.S, Dealwis, C. | | 登録日 | 2008-04-09 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase: A Conformationally Flexible Pharmacophore
J.Med.Chem., 51, 2008
|
|
2YN4
 
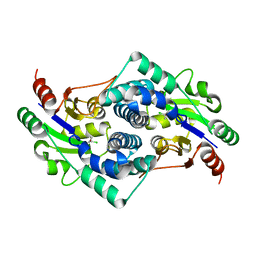 | | L-2-chlorobutryic acid bound complex L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | 分子名称: | (2S)-2-chlorobutanoic acid, L-HALOACID DEHALOGENASE | | 著者 | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | 登録日 | 2012-10-12 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMP
 
 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | 分子名称: | L-HALOACID DEHALOGENASE | | 著者 | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | 登録日 | 2012-10-10 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YML
 
 | | Native L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | 分子名称: | L-HALOACID DEHALOGENASE | | 著者 | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | 登録日 | 2012-10-09 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMQ
 
 | | Chloropropionic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | 分子名称: | L-HALOACID DEHALOGENASE | | 著者 | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | 登録日 | 2012-10-10 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
3AML
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | 登録日 | 2010-08-20 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AMK
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | 分子名称: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | 著者 | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | 登録日 | 2010-08-20 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
