3LKU
 
 | |
2GOY
 
 | | Crystal structure of assimilatory adenosine 5'-phosphosulfate reductase with bound APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, IRON/SULFUR CLUSTER, adenosine phosphosulfate reductase | | Authors: | Chartron, J, Carroll, K.S, Shiau, C, Gao, H, Leary, J.A, Bertozzi, C.R, Stout, C.D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate Recognition, Protein Dynamics, and Iron-Sulfur Cluster in Pseudomonas aeruginosa Adenosine 5'-Phosphosulfate Reductase.
J.Mol.Biol., 364, 2006
|
|
3VEJ
 
 | | Crystal structure of the Get5 carboxyl domain from S. cerevisiae | | Descriptor: | PHOSPHATE ION, Ubiquitin-like protein MDY2 | | Authors: | Chartron, J.W, Vandervelde, D.G, Rao, M, Clemons Jr, W.M. | | Deposit date: | 2012-01-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Get5 Carboxyl-terminal Domain Is a Novel Dimerization Motif That Tethers an Extended Get4/Get5 Complex.
J.Biol.Chem., 287, 2012
|
|
3SZ7
 
 | |
4GOE
 
 | |
4GOF
 
 | |
4GOD
 
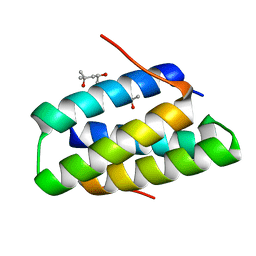 | | Crystal structure of the SGTA homodimerization domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Small glutamine-rich tetratricopeptide repeat-containing protein alpha | | Authors: | Chartron, J.W, VanderVelde, D.G, Clemons Jr, W.M. | | Deposit date: | 2012-08-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of the Sgt2/SGTA Dimerization Domain with the Get5/UBL4A UBL Domain Reveal an Interaction that Forms a Conserved Dynamic Interface.
Cell Rep, 2, 2012
|
|
4GOC
 
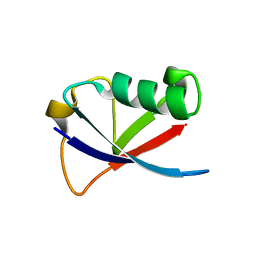 | |
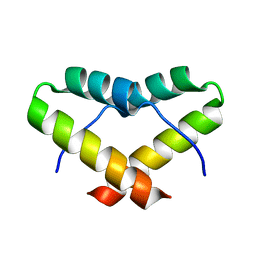
2LNZ
 
 | |
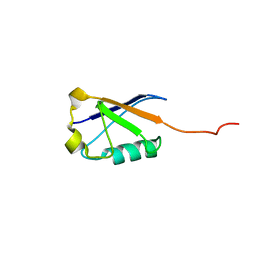
2LXA
 
 | |
2LXB
 
 | |
2LXC
 
 | |
2O8V
 
 | | PAPS reductase in a covalent complex with thioredoxin C35A | | Descriptor: | Phosphoadenosine phosphosulfate reductase, Thioredoxin 1 | | Authors: | Chartron, J, Shiau, C, Stout, C.D, Carroll, K.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Phosphoadenosine-5'-phosphosulfate Reductase in Complex with Thioredoxin: A Structural Snapshot in the Catalytic Cycle.
Biochemistry, 46, 2007
|
|
2LO0
 
 | |
2OG2
 
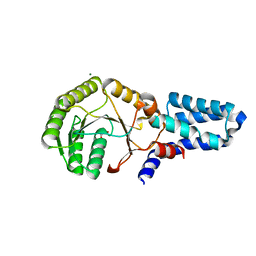 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
4WWR
 
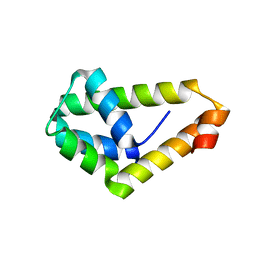 | |
1XLT
 
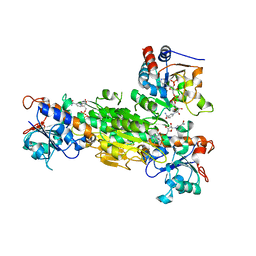 | | Crystal structure of Transhydrogenase [(domain I)2:domain III] heterotrimer complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sundaresan, V, Chartron, J, Yamaguchi, M, Stout, C.D. | | Deposit date: | 2004-09-30 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational Diversity in NAD(H) and Transhydrogenase Nicotinamide Nucleotide Binding Domains
J.Mol.Biol., 346, 2005
|
|
5BW8
 
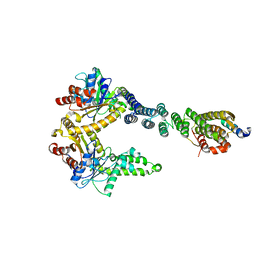 | | 2.8 A crystal structure of a Get3-Get4-Get5 intermediate complex from S.cerevisiae | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 4, Ubiquitin-like protein MDY2, ... | | Authors: | Gristick, H.B, Chartron, J.W, Clemons, W.M. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting.
J.Biol.Chem., 290, 2015
|
|
5BWK
 
 | | 6.0 A Crystal structure of a Get3-Get4-Get5 intermediate complex from S.cerevisiae | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 4, Ubiquitin-like protein MDY2, ... | | Authors: | Gristick, H.B, Chartron, J.W, Clemons, W.M. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting.
J.Biol.Chem., 290, 2015
|
|
5W74
 
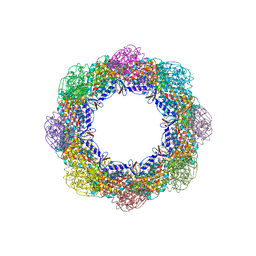 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis D386ADeltaLid Mutant in the Open, ADP-Bound State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
5W79
 
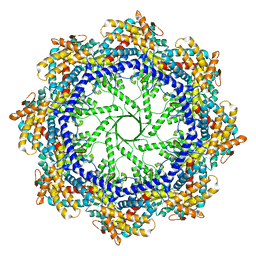 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis, Cysteine-less mutant in the Apo State | | Descriptor: | Chaperonin, SULFATE ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (3.122 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
3IDQ
 
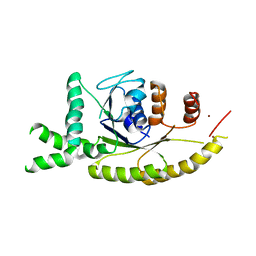 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IBG
 
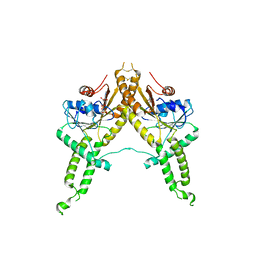 | | Crystal structure of Aspergillus fumigatus Get3 with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase, subunit of the Get complex | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1PNO
 
 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADP | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PNQ
 
 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADPH | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
