7Q9E
 
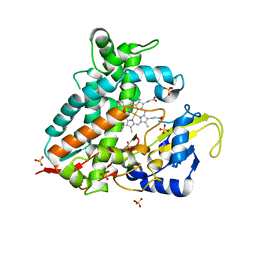 | | CYP106A1 | | Descriptor: | COBALT (II) ION, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carius, Y, Hutter, M, Kiss, F, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of the cytochrome P450 enzymes CYP106A1 and CYP106A2 provides insight into their differences in steroid conversion.
Febs Lett., 596, 2022
|
|
2WDZ
 
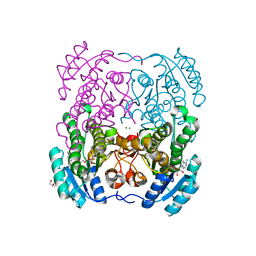 | | Crystal structure of the short chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD+ and 1,2-Pentandiol | | Descriptor: | (2S)-pentane-1,2-diol, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
7ZGI
 
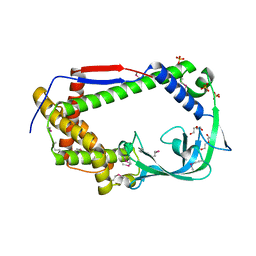 | | chloroplast trigger factor (TIG1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidylprolyl isomerase, SULFATE ION | | Authors: | Carius, Y, Ries, F, Gries, K, Trentmann, O, Willmund, F, Lancaster, C.R.D. | | Deposit date: | 2022-04-03 | | Release date: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features of chloroplast trigger factor determined at 2.6 angstrom resolution.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3DML
 
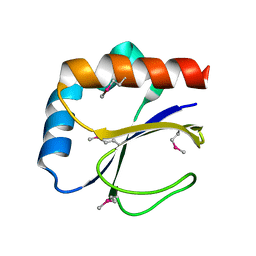 | |
3D4T
 
 | |
7ZZL
 
 | | Crystal structure of CYP106A1 | | Descriptor: | COBALT (II) ION, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carius, Y, Kiss, F, Hutter, M, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of the cytochrome P450 enzymes CYP106A1 and CYP106A2 provides insight into their differences in steroid conversion.
Febs Lett., 596, 2022
|
|
2WSB
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD | | Descriptor: | GALACTITOL DEHYDROGENASE, MAGNESIUM ION, N-PROPANOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-09-04 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
5LIV
 
 | | Crystal structure of myxobacterial CYP260A1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 CYP260A1,Cytochrome P450 CYP260A1, DIMETHYL SULFOXIDE, ... | | Authors: | Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of CYP260A1 from Sorangium cellulosum to investigate the 1 alpha-hydroxylation of a mineralocorticoid.
FEBS Lett., 590, 2016
|
|
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | Descriptor: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
4YT3
 
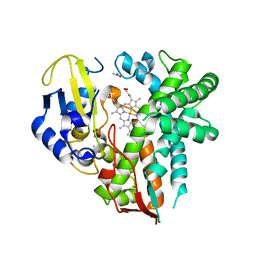 | | CYP106A2 | | Descriptor: | ACETATE ION, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | janocha, S, carius, y, bernhardt, r, lancaster, c.r.d. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
4P1Q
 
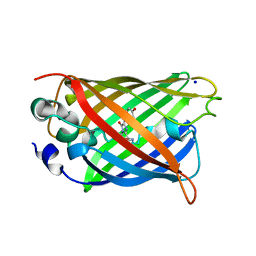 | | GREEN FLUORESCENT PROTEIN E222H VARIANT | | Descriptor: | Green fluorescent protein, SODIUM ION | | Authors: | Klein, M, Carius, Y, Auerbach, D, Franz, S, Jung, G, Lancaster, C.R.D. | | Deposit date: | 2014-02-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Replacement of Highly Conserved E222 by the Photostable Non-photoconvertible Histidine in GFP.
Chembiochem, 15, 2014
|
|
5IKI
 
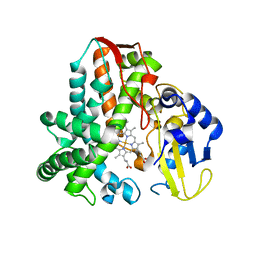 | | CYP106A2 WITH SUBSTRATE ABIETIC ACID | | Descriptor: | Abietic acid, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Janocha, S, Carius, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
2YKT
 
 | | Crystal structure of the I-BAR domain of IRSp53 (BAIAP2) in complex with an EHEC derived Tir peptide | | Descriptor: | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 1-ASSOCIATED PROTEIN 2, SULFATE ION, TRANSLOCATED INTIMIN RECEPTOR PROTEIN | | Authors: | de Groot, J.C, Schlueter, K, Carius, Y, Quedenau, C, Vingadassalom, D, Faix, J, Weiss, S.M, Reichelt, J, Standfuss-Gabisch, C, Lesser, C.F, Leong, J.M, Heinz, D.W, Buessow, K, Stradal, T.E.B. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
Structure, 19, 2011
|
|
5HIW
 
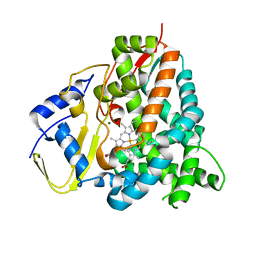 | | Sorangium cellulosum So Ce56 cytochrome P450 260B1 | | Descriptor: | Cytochrome P450 CYP260B1, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Salamanca-Pinzon, S.G, Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-01-12 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis for the hydroxylation of Delta 4 C21-steroids by the myxobacterial CYP260B1.
Febs Lett., 590, 2016
|
|
3NHB
 
 | | Nucleotide Binding Domain of Human ABCB6 (ADP bound structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NHA
 
 | | Nucleotide Binding Domain of Human ABCB6 (ADP Mg bound structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NH9
 
 | | Nucleotide Binding Domain of Human ABCB6 (ATP bound structure) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NH6
 
 | | Nucleotide Binding Domain of human ABCB6 (apo structure) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, BETA-MERCAPTOETHANOL | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
