4J9C
 
 | |
4J9G
 
 | |
4JJD
 
 | |
4J9I
 
 | |
4J9F
 
 | |
4JJC
 
 | |
1JGZ
 
 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Lys | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JH0
 
 | | Photosynthetic Reaction Center Mutant With Glu L 205 Replaced to Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGW
 
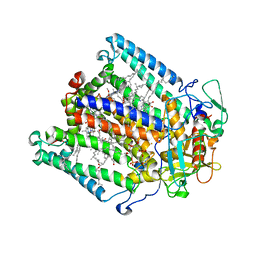 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JDL
 
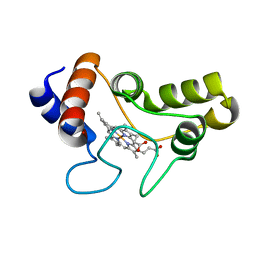 | | Structure of cytochrome c2 from Rhodospirillum Centenum | | Descriptor: | CYTOCHROME C2, ISO-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Camara-Artigas, A, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-14 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of cytochrome c2 from Rhodospirillum centenum.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGY
 
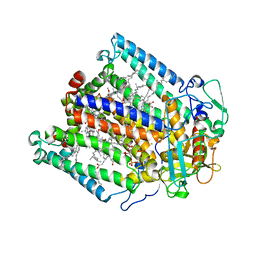 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Phe | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGX
 
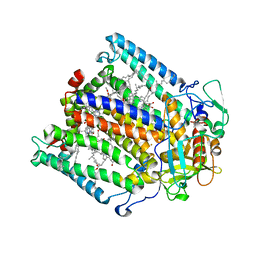 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Asp | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3I9Q
 
 | |
4QT7
 
 | |
3HG6
 
 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | Descriptor: | GLYCEROL, Onconase, SULFATE ION | | Authors: | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
7OL8
 
 | |
7OL6
 
 | |
7OL7
 
 | |
7OL5
 
 | |
4OMN
 
 | |
4OMQ
 
 | |
4OMM
 
 | |
3M0P
 
 | |
3M0S
 
 | |
3M0T
 
 | |
