5N5N
 
 | | Cryo-EM structure of tsA201 cell alpha1B and betaI and betaIVb microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Vemu, A, Atherton, J, Spector, J.O, Moores, C.A, Roll-Mecak, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tubulin isoform composition tunes microtubule dynamics.
Mol. Biol. Cell, 28, 2017
|
|
6G4A
 
 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
5ND2
 
 | | Microtubule-bound MKLP2 motor domain in the presence of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Yu, I.M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
6G00
 
 | | Crystal Structure of a GH8 xylanase from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, SODIUM ION | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5ND4
 
 | | Microtubule-bound MKLP2 motor domain in the presence of ADP.AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Yu, I.-M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
8SF5
 
 | |
8SF6
 
 | | Promiscuous amino acid gamma synthase from Caldicellulosiruptor hydrothermalis in closed conformation | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Buller, A.R, Zmich, A.P, Bingman, C.A. | | Deposit date: | 2023-04-10 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Multiplexed Assessment of Promiscuous Non-Canonical Amino Acid Synthase Activity in a Pyridoxal Phosphate-Dependent Protein Family.
Acs Catalysis, 13, 2023
|
|
5ND3
 
 | | Microtubule-bound MKLP2 motor domain in the with no nucleotide | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF20A, ... | | Authors: | Atherton, J, Yu, I.M, Cook, A, Muretta, J.M, Joseph, A.P, Major, J, Sourigues, Y, Clause, J, Topf, M, Rosenfeld, S.S, Houdusse, A, Moores, C.A. | | Deposit date: | 2017-03-07 | | Release date: | 2017-10-04 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry.
Elife, 6, 2017
|
|
8SVC
 
 | | Crystal Structure of SBP from Klebsiella pneumoniae | | Descriptor: | Metal ABC transporter substrate-binding protein, SULFATE ION, ZINC ION | | Authors: | Giles, M.W, Cole, G.B, Ng, D, McDevitt, C.A, Moraes, T.F. | | Deposit date: | 2023-05-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Zinc acquisition and its contribution to Klebsiella pneumoniae virulence.
Front Cell Infect Microbiol, 13, 2023
|
|
5NMS
 
 | | Hsp21 dodecamer, structural model based on cryo-EM and homology modelling | | Descriptor: | 25.3 kDa heat shock protein, chloroplastic | | Authors: | Rutsdottir, G, Harmark, J, Koeck, P.J.B, Hebert, H, Soderberg, C.A.G, Emanuelsson, C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity.
J. Biol. Chem., 292, 2017
|
|
7N2T
 
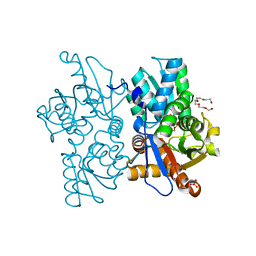 | | O-acetylserine sulfhydrylase from Citrullus vulgaris in the internal aldimine state, with citrate bound | | Descriptor: | CITRIC ACID, Cysteine synthase, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, J.L, Buller, A.R, Bingman, C.A. | | Deposit date: | 2021-05-29 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of beta-Substitution Activity of O-Acetylserine Sulfhydrolase from Citrullus vulgaris.
Chembiochem, 23, 2022
|
|
5M54
 
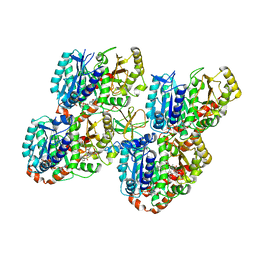 | | Mechanism of microtubule minus-end recognition and protection by CAMSAP proteins | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Akhmanova, A, Moores, C.A, Baldus, M, Steinmetz, M.O, Topf, M, Roberts, A.J, Grant, B.J, Scarabelli, G, Joseph, A.-J, van Hooff, J.J.E, Houben, K, Hua, S, Luo, Y, Stangier, M.M, Jiang, K, Atherton, J. | | Deposit date: | 2016-10-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A structural model for microtubule minus-end recognition and protection by CAMSAP proteins.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7ZGH
 
 | |
7ZGG
 
 | |
7ZGF
 
 | |
6G81
 
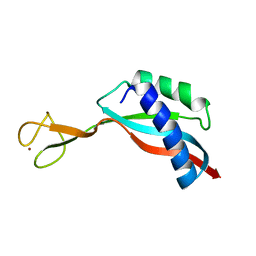 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | Descriptor: | Hydrogenase maturation factor HypA, ZINC ION | | Authors: | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | Deposit date: | 2018-04-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
7O19
 
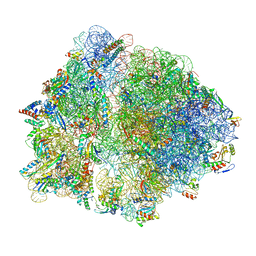 | | Cryo-EM structure of an Escherichia coli TnaC-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1C
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome-RF2 complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1A
 
 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
5MPT
 
 | | Structure of the citrinin polyketide synthase CMeT domain | | Descriptor: | 1,2-ETHANEDIOL, Citrinin polyketide synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Herbst, D.A, Storm, P.A, Townsend, C.A, Maier, T. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Functional and Structural Analysis of Programmed C-Methylation in the Biosynthesis of the Fungal Polyketide Citrinin.
Cell Chem Biol, 24, 2017
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GO4
 
 | | TdT chimera (Loop1 of pol mu) - binary complex with ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase, MAGNESIUM ION, ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GO3
 
 | | TdT chimera (Loop1 of pol mu) - apoenzyme | | Descriptor: | DNA nucleotidylexotransferase,DNA-directed DNA/RNA polymerase mu,DNA nucleotidylexotransferase | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6GJ9
 
 | |
