4LUF
 
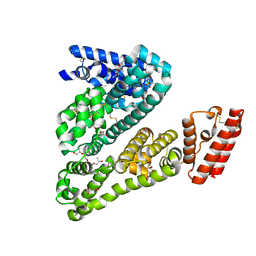 | | Crystal Structure of Ovine Serum Albumin | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, (2S)-2-hydroxybutanedioic acid, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J.A, Pietrzyk, A.J, Bujacz, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of ovine serum albumin and its complex with 3,5-diiodosalicylic acid
To be Published
|
|
4RKD
 
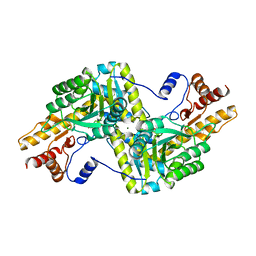 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with aspartic acid | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYLENE)-AMINO]-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aromatic amino acid aminotransferase, ... | | Authors: | Bujacz, A, Rutkiewicz-Krotewicz, M, Bujacz, G, Nowakowska-Sapota, K, Turkiewicz, M. | | Deposit date: | 2014-10-12 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure and enzymatic properties of a broad substrate-specificity psychrophilic aminotransferase from the Antarctic soil bacterium Psychrobacter sp. B6.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RKC
 
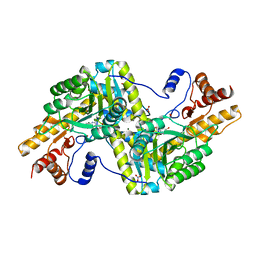 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aromatic amino acid aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rutkiewicz-Krotewicz, M, Bujacz, G, Nowakowska-Sapota, K, Turkiewicz, M. | | Deposit date: | 2014-10-12 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure and enzymatic properties of a broad substrate-specificity psychrophilic aminotransferase from the Antarctic soil bacterium Psychrobacter sp. B6.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
3PIG
 
 | | beta-fructofuranosidase from Bifidobacterium longum | | Descriptor: | Beta-fructofuranosidase, CHLORIDE ION | | Authors: | Bujacz, A, Bujacz, G, Redzynia, I, Krzepkowska-Jedrzejczak, M, Bielecki, S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the apo form of beta-fructofuranosidase from Bifidobacterium longum and its complex with fructose
Febs J., 278, 2011
|
|
3PIJ
 
 | | beta-fructofuranosidase from Bifidobacterium longum - complex with fructose | | Descriptor: | Beta-fructofuranosidase, CHLORIDE ION, beta-D-fructofuranose | | Authors: | Bujacz, A, Bujacz, G, Redzynia, I, Krzepkowska-Jedrzejczak, M, Bielecki, S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the apo form of beta-fructofuranosidase from Bifidobacterium longum and its complex with fructose
Febs J., 278, 2011
|
|
8BSG
 
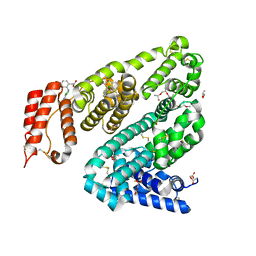 | | COMPLEX OF LEPORINE SERUM ALBUMIN WITH DICLOFENAC | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Bujacz, A, Talaj, J, Zielinski, K. | | Deposit date: | 2022-11-25 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
4GLF
 
 | |
4GLJ
 
 | | Crystal structure of methylthioadenosine phosphorylase in complex with rhodamine B | | Descriptor: | CHLORIDE ION, N-[9-(2-carboxyphenyl)-6-(diethylamino)-3H-xanthen-3-ylidene]-N-ethylethanaminium, PHOSPHATE ION, ... | | Authors: | Bujacz, A, Bujacz, G, Cieslinski, H, Bartasun, P. | | Deposit date: | 2012-08-14 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A study on the interaction of rhodamine B with methylthioadenosine phosphorylase protein sourced from an antarctic soil metagenomic library.
Plos One, 8, 2013
|
|
6ZUP
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
4F5S
 
 | | Crystal Structure of Bovine Serum Albumin | | Descriptor: | Serum albumin, TRIETHYLENE GLYCOL | | Authors: | Bujacz, A, Bujacz, G. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of bovine, equine and leporine serum albumin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F5V
 
 | | Crystal Structure of Leporine Serum Albumin | | Descriptor: | ACETATE ION, Serum albumin, TETRAETHYLENE GLYCOL, ... | | Authors: | Bujacz, A, Bujacz, G. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of bovine, equine and leporine serum albumin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F5T
 
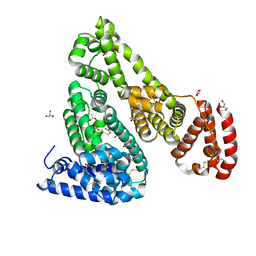 | | Crystal Structure of Equine Serum Albumin | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Bujacz, A, Bujacz, G. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of bovine, equine and leporine serum albumin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F5U
 
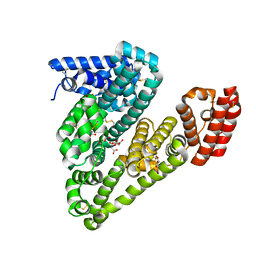 | | Crystal structure of Equine Serum Albumin at 2.04 resolution | | Descriptor: | (2S)-2-hydroxybutanedioic acid, MALONATE ION, SUCCINIC ACID, ... | | Authors: | Bujacz, A, Bujacz, G. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of bovine, equine and leporine serum albumin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4LUH
 
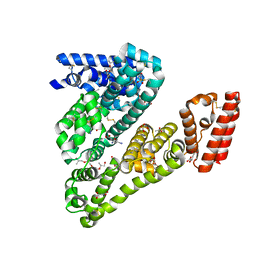 | | Complex of ovine serum albumin with 3,5-diiodosalicylic acid | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, (2S)-2-hydroxybutanedioic acid, 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, ... | | Authors: | Bujacz, A, Talaj, J.A, Pietrzyk, A.J, Bujacz, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ovine serum albumin and its complex with 3,5-diiodosalicylic acid
To be Published
|
|
5ORI
 
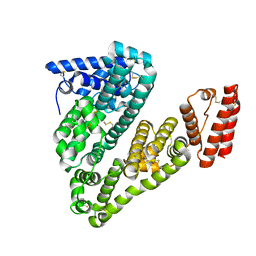 | | Structure of caprine serum albumin in orthorhombic crystal system | | Descriptor: | Albumin | | Authors: | Bujacz, A, Talaj, J.A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6ETZ
 
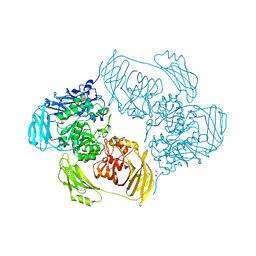 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted Beta-D-Galactosidases: Crystal Structure of BetaDG from Arthrobacter sp. 32cB
Crystals, 8, 2018
|
|
5OSW
 
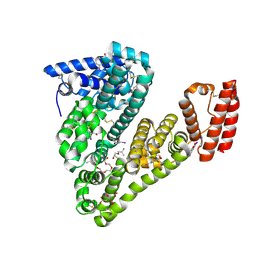 | | Structure of caprine serum albumin in complex with 3,5-diiodosalicylic acid | | Descriptor: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LDR
 
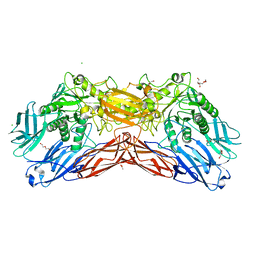 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain in complex with galactose | | Descriptor: | ACETATE ION, Beta-D-galactosidase, CHLORIDE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
6SED
 
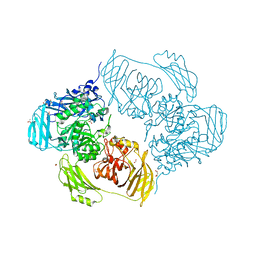 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SEB
 
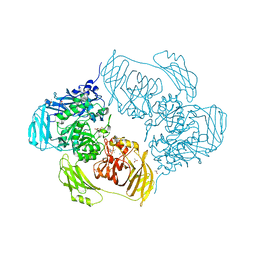 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
5EUV
 
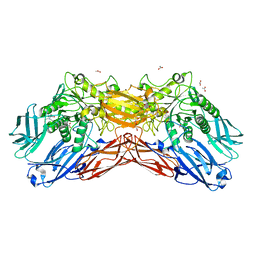 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6H1P
 
 | |
