6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
3HU9
 
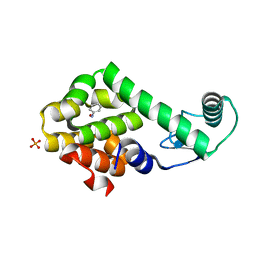 | | Nitrosobenzene in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, NITROSOBENZENE, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT8
 
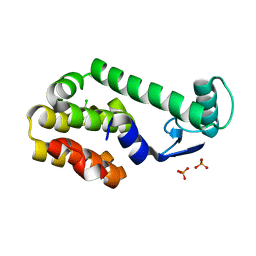 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT6
 
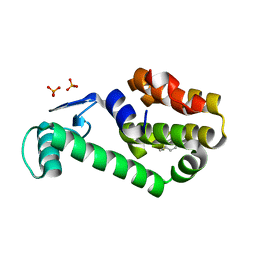 | | 2-methylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, PHOSPHATE ION, o-cresol | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HU8
 
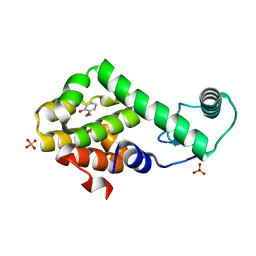 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT7
 
 | | 2-ethylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTF
 
 | | 4-chloro-1h-pyrazole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4-chloro-1H-pyrazole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HT9
 
 | | 2-methoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Guaiacol, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUQ
 
 | | Thieno[3,2-b]thiophene in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUK
 
 | | Benzylacetate in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTB
 
 | | 2-propylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-propylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTD
 
 | | (Z)-Thiophene-2-carboxaldoxime in complex with T4 lysozyme L99A/M102Q | | Descriptor: | (NZ)-N-(thiophen-2-ylmethylidene)hydroxylamine, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
4JPU
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with Benzamidine | | Descriptor: | BENZAMIDINE, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
5AK0
 
 | | Human PFKFB3 in complex with an indole inhibitor 6 | | Descriptor: | (2S)-N-[4-[1-METHYL-3-(1-METHYLPYRAZOL-4-YL)INDOL-5-YL]OXYPHENYL]PYRROLIDINE-2-CARBOXAMIDE, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5AJX
 
 | | Human PFKFB3 in complex with an indole inhibitor 3 | | Descriptor: | (2S)-N-[4-[3-cyano-1-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]indol-5-yl]oxyphenyl]pyrrolidine-2-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
4JQM
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinazoline | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
5AJZ
 
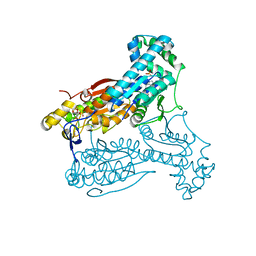 | | Human PFKFB3 in complex with an indole inhibitor 5 | | Descriptor: | 2-azanyl-N-[4-[(3-cyano-1H-indol-5-yl)oxy]phenyl]ethanamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5AJW
 
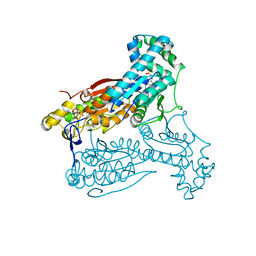 | | Human PFKFB3 in complex with an indole inhibitor 2 | | Descriptor: | 2-amino-N-[4-(2-amino-1-benzyl-3-cyano-indol-5-yl)oxyphenyl]acetamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
4JPT
 
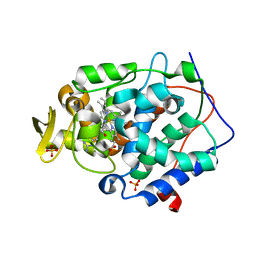 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with quinazoline-2,4-diamine | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
5AJY
 
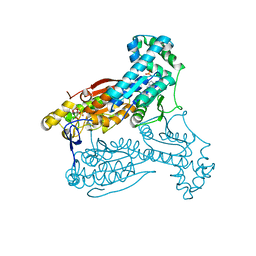 | | Human PFKFB3 in complex with an indole inhibitor 4 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, N-(4-{[3-(1-methyl-1H-pyrazol-4-yl)-1H-indol-5-yl]oxy}phenyl)glycinamide, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
4JQJ
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinoline | | Descriptor: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-20 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
5AJV
 
 | | Human PFKFB3 in complex with an indole inhibitor 1 | | Descriptor: | (2S)-2-amino-N-[4-[(2-amino-3-cyano-1H-indol-5-yl)oxy]phenyl]-3-hydroxy-propanamide, 2,6-di-O-phosphono-beta-D-fructofuranose, HUMAN PFKFB3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
4JPL
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Azaindole | | Descriptor: | 1H-pyrrolo[3,2-b]pyridine, Cytochrome c peroxidase, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
