8TC1
 
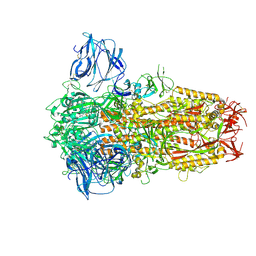 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A.R. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC0
 
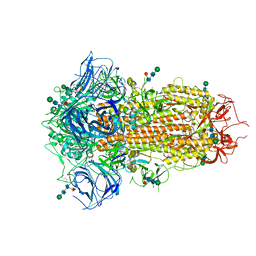 | | Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC5
 
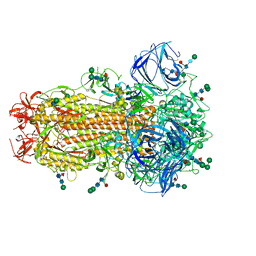 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8CXP
 
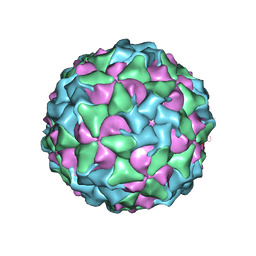 | | Characterisation of a Seneca Valley Virus Thermostable Mutant | | Descriptor: | Capsid protein VP1, Capsid protein VP3, VP2, ... | | Authors: | Jayawardena, N, Bostina, M, Strauss, M. | | Deposit date: | 2022-05-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Characterisation of a Seneca Valley virus thermostable mutant.
Virology, 575, 2022
|
|
7THX
 
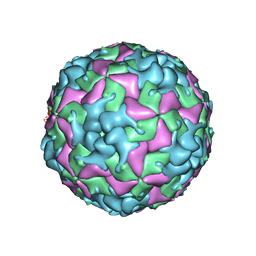 | | Cryo-EM structure of W6 possum enterovirus | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, I, Jayawardena, N, Strauss, M, Bostina, M. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM Structure of a Possum Enterovirus.
Viruses, 14, 2022
|
|
6CX1
 
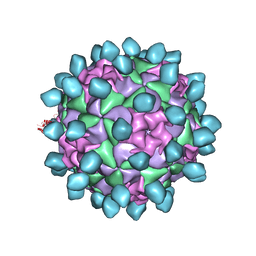 | | Cryo-EM structure of Seneca Valley Virus-Anthrax Toxin Receptor 1 complex | | Descriptor: | Anthrax toxin receptor 1, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Jayawardena, N, Burga, L, Easingwood, R, Takizawa, Y, Wolf, M, Bostina, M. | | Deposit date: | 2018-04-02 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3IYB
 
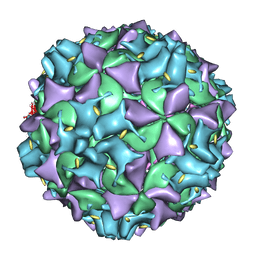 | | Poliovirus early RNA-release intermediate | | Descriptor: | Genome polyprotein, Precursor polyprotein, VP1 core | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
3IYC
 
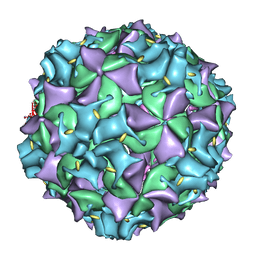 | | Poliovirus late RNA-release intermediate | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Genome polyprotein, ... | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
