2Q60
 
 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
3ZGN
 
 | | Crystal structures of Escherichia coli IspH in complex with TMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
2P0U
 
 | | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2) | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Borel, F, Schroeder, G, Schroeder, J, Ferrer, J.-L. | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | crystal structure of Marchantia polymorpha stilbenecarboxylate synthase 2 (STCS2)
To be Published
|
|
4BV8
 
 | | Crystal structure of the apo form of mouse Mu-crystallin. | | Descriptor: | GLYCEROL, POTASSIUM ION, THIOMORPHOLINE-CARBOXYLATE DEHYDROGENASE | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
4BV9
 
 | | Crystal structure of the NADPH form of mouse Mu-crystallin. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
4BVA
 
 | | Crystal structure of the NADPH-T3 form of mouse Mu-crystallin. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Borel, F, Hachi, I, Palencia, A, Gaillard, M.C, Ferrer, J.L. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mouse Mu-Crystallin Complexed with Nadph and the T3 Thyroid Hormone
FEBS J., 281, 2014
|
|
3HJU
 
 | | Crystal structure of human monoglyceride lipase | | Descriptor: | GLYCEROL, Monoglyceride lipase | | Authors: | Labar, G, Bauvois, C, Borel, F, Ferrer, J.-L, Wouters, J, Lambert, D.M. | | Deposit date: | 2009-05-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human Monoacylglycerol Lipase, a Key Actor in Endocannabinoid Signaling
Chembiochem, 11, 2010
|
|
3CIA
 
 | | Crystal structure of cold-aminopeptidase from Colwellia psychrerythraea | | Descriptor: | ZINC ION, cold-active aminopeptidase | | Authors: | Bauvois, C, Jacquamet, L, Borel, F, Ferrer, J.-L. | | Deposit date: | 2008-03-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Cold-active Aminopeptidase from Colwellia psychrerythraea, a Close Structural Homologue of the Human Bifunctional Leukotriene A4 Hydrolase.
J.Biol.Chem., 283, 2008
|
|
1XIU
 
 | | Crystal structure of the agonist-bound ligand-binding domain of Biomphalaria glabrata RXR | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 1, RXR-like protein | | Authors: | De Groot, A, De Rosny, E, Juillan-Binard, C, Ferrer, J.-L, Laudet, V, Pebay-Peroula, E, Fontecilla-Camps, J.-C, Borel, F. | | Deposit date: | 2004-09-22 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Novel Tetrameric Complex of Agonist-bound Ligand-binding Domain of Biomphalaria glabrata Retinoid X Receptor.
J.Mol.Biol., 354, 2005
|
|
2BKW
 
 | | Yeast alanine:glyoxylate aminotransferase YFL030w | | Descriptor: | ALANINE-GLYOXYLATE AMINOTRANSFERASE 1, GLYOXYLIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meyer, P, Liger, D, Leulliot, N, Quevillon-Cheruel, S, Zhou, C.Z, Borel, F, Ferrer, J.L, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2005-02-21 | | Release date: | 2005-11-02 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure and Confirmation of the Alanine:Glyoxylate Aminotransferase Activity of the Yfl030W Yeast Protein
Biochimie, 87, 2005
|
|
4MMO
 
 | | The crystal structure of a M20 family metallo-carboxypeptidase Sso-CP2 from Sulfolobus solfataricus | | Descriptor: | GLYCEROL, SULFATE ION, Sso-CP2 metallo-carboxypetidase, ... | | Authors: | Dupuy, J, Dutoit, R, Durisotti, V, Demarez, M, Borel, F, Van Elder, D, Legrain, C, Bauvois, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3363 Å) | | Cite: | Biochemical characterization of a novel thermostable dinuclear carboxypeptidase from the thermoacidophilic archaeum Sulfolobus solfataricus.
To be Published
|
|
1TJ4
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ5
 
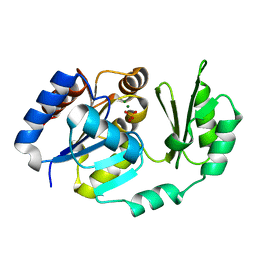 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose and phosphate | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Sucrose-Phosphatase, ... | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1U2T
 
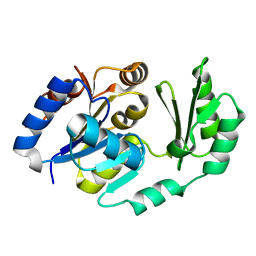 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose6P | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, sucrose-phosphatase (SPP) | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
1U2S
 
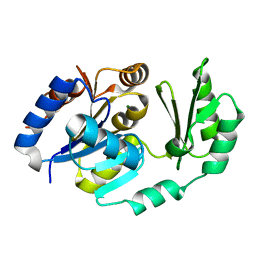 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with glucose | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
1S2O
 
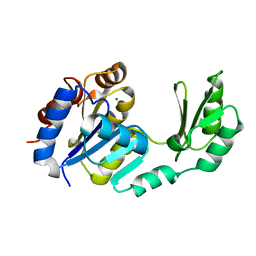 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | | Descriptor: | MAGNESIUM ION, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.L. | | Deposit date: | 2004-01-09 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ3
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in a closed conformation | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
