3T4B
 
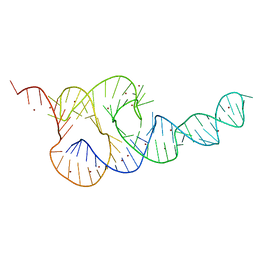 | | Crystal Structure of the HCV IRES pseudoknot domain | | Descriptor: | HCV IRES pseudoknot domain plus crystallization module, NICKEL (II) ION | | Authors: | Berry, K.E, Waghray, S, Mortimer, S.A, Bai, Y, Doudna, J.A. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning.
Structure, 19, 2011
|
|
3IHQ
 
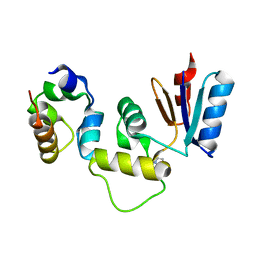 | |
3D71
 
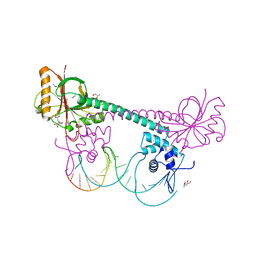 | | Crystal structure of E253Q BMRR bound to 22 base pair promoter site | | Descriptor: | BMR promoter DNA, CITRATE ANION, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Huffman, J.L, Brennan, R.G. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of BmrR-Drug Complexes Reveal a Rigid Multidrug Binding Pocket and Transcription Activation through Tyrosine Expulsion
J.Biol.Chem., 283, 2008
|
|
3D70
 
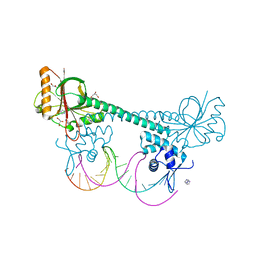 | |
1R8E
 
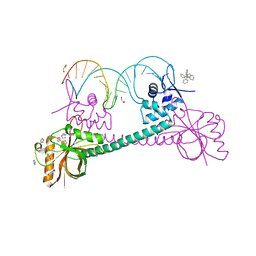 | | Crystal Structure of BmrR Bound to DNA at 2.4A Resolution | | Descriptor: | 5'-D(*GP*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*TP*C)-3', GLYCEROL, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
3D6Y
 
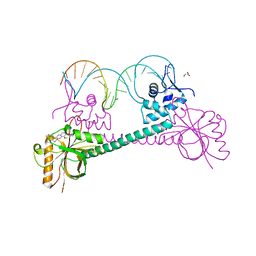 | |
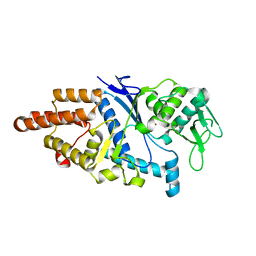
1LI5
 
 | |
1LI7
 
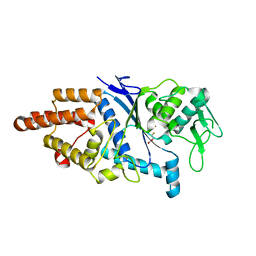 | |
3D6Z
 
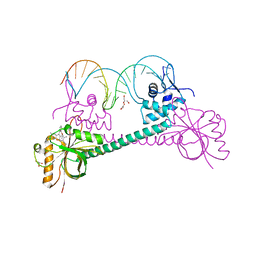 | |
1R8D
 
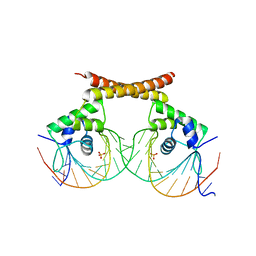 | | Crystal Structure of MtaN Bound to DNA | | Descriptor: | 26-MER, SULFATE ION, transcription activator MtaN | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
1Z3E
 
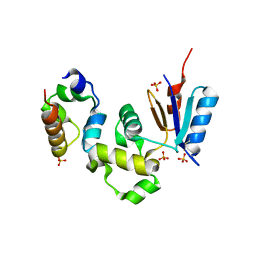 | | Crystal Structure of Spx in Complex with the C-terminal Domain of the RNA Polymerase Alpha Subunit | | Descriptor: | DNA-directed RNA polymerase alpha chain, Regulatory protein spx, SULFATE ION | | Authors: | Newberry, K.J, Nakano, S, Zuber, P, Brennan, R.G. | | Deposit date: | 2005-03-11 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the {alpha} C-terminal domain of RNA polymerase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6PYO
 
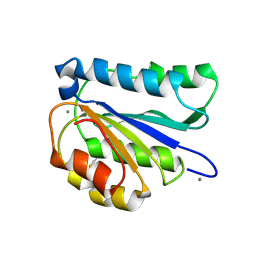 | |
6PYX
 
 | |
4J1R
 
 | | Crystal Structure of GSK3b in complex with inhibitor 15R | | Descriptor: | (2R)-2-(1H-indol-3-ylmethyl)-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-01 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
4J71
 
 | | Crystal Structure of GSK3b in complex with inhibitor 1R | | Descriptor: | (2R)-2-methyl-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, CHLORIDE ION, Glycogen synthase kinase-3 beta, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-12 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
3IAO
 
 | |
2PEX
 
 | |
1EUY
 
 | | GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA MUTANT AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-19 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
1EUQ
 
 | | CRYSTAL STRUCTURE OF GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA-GLN MUTANT AND AN ACTIVE-SITE INHIBITOR | | Descriptor: | 5'-O-[N-(L-GLUTAMINYL)-SULFAMOYL]ADENOSINE, GLUTAMINYL TRNA, GLUTAMINYL-TRNA SYNTHETASE | | Authors: | Sherlin, L.D, Bullock, T.L, Newberry, K.J, Lipman, R.S.A, Hou, Y.-M, Beijer, B, Sproat, B.S, Perona, J.J. | | Deposit date: | 2000-04-17 | | Release date: | 2000-06-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.
J.Mol.Biol., 299, 2000
|
|
2PFB
 
 | |
