3BRL
 
 | |
3E2B
 
 | | Crystal structure of Dynein Light chain LC8 in complex with a peptide derived from Swallow | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Barbar, E, Karplus, P.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interplay of ligand binding and quaternary structure in the diverse interactions of dynein light chain LC8.
J.Mol.Biol., 384, 2008
|
|
2P2T
 
 | | Crystal structure of dynein light chain LC8 bound to residues 123-138 of intermediate chain IC74 | | Descriptor: | ACETATE ION, Dynein intermediate chain peptide, Dynein light chain 1, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E. | | Deposit date: | 2007-03-07 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site.
J.Mol.Biol., 371, 2007
|
|
3BRI
 
 | | Crystal Structure of apo-LC8 | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E, Chiodo, M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interplay of Ligand Binding and Quaternary Structure in the Diverse Interactions of Dynein Light Chain LC8.
J.Mol.Biol., 384, 2008
|
|
1S6L
 
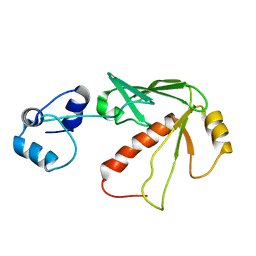 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
