1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
3ID0
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with 3-Fluoro-1-(2-hydroxy-2,2-bisphosphono-ethyl)pyridinium | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-19 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3O6Z
 
 | | Structure of the D152A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O69
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GDP-mannose pyrophosphatase nudK, ... | | Authors: | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | Deposit date: | 2010-07-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3Q4I
 
 | | Crystal structure of CDP-Chase in complex with Gd3+ | | Descriptor: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
3Q1P
 
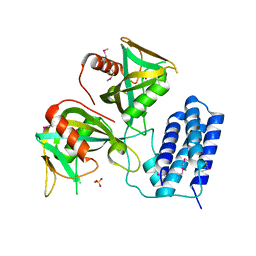 | | Crystal structure of CDP-Chase | | Descriptor: | Phosphohydrolase (MutT/nudix family protein), SULFATE ION | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-17 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
3HIZ
 
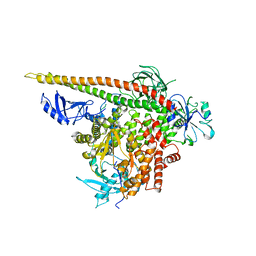 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HHM
 
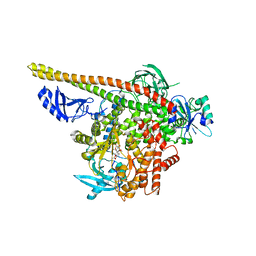 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ICZ
 
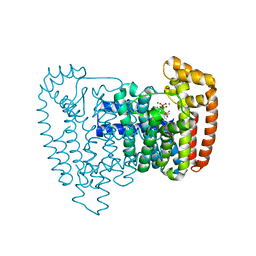 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with 1-(2,2-Bis-phosphono-ethyl)-3-butyl-pyridinium and isopentenyl pyrophosphate | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 3-[(1E)-but-1-en-1-yl]-1-(2,2-diphosphonoethyl)pyridinium, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-19 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3JBJ
 
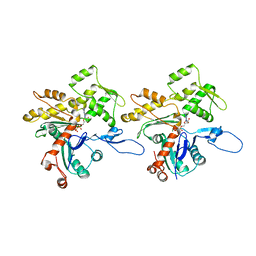 | | Cryo-EM reconstruction of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
1NQY
 
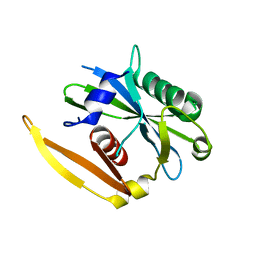 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NQZ
 
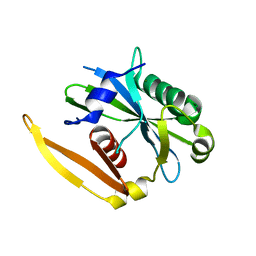 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
