7A1G
 
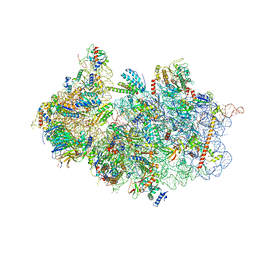 | | Structure of a crosslinked yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Mackens-Kiani, T, Kratzat, H, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7AFT
 
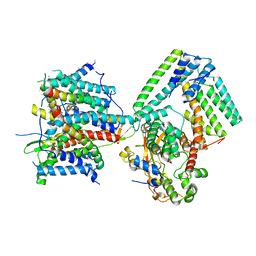 | | Cryo-EM structure of the signal sequence-engaged post-translational Sec translocon | | Descriptor: | Mating factor alpha-1,Mating factor alpha-1, Protein translocation protein SEC63, Protein transport protein SBH1, ... | | Authors: | Weng, T.-H, Beatrix, B, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the active post-translational Sec translocon.
Embo J., 40, 2021
|
|
7Z20
 
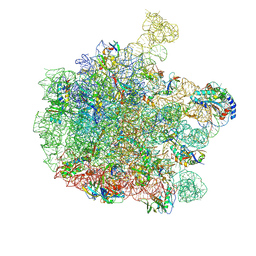 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
1RY1
 
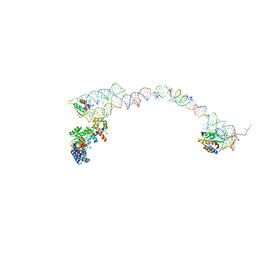 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | Descriptor: | SRP Alu domain, SRP RNA, SRP S domain, ... | | Authors: | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
7ZS5
 
 | | Structure of 60S ribosomal subunit from S. cerevisiae with eIF6 and tRNA | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZRS
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUX
 
 | | Collided ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZPQ
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUW
 
 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZP8
 
 | | 70S E. coli ribosome with a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Chan, S.H.S, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ5
 
 | | 70S E. coli ribosome with truncated uL23 and uL24 loops | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ6
 
 | | 70S E. coli ribosome with truncated uL23 and uL24 loops and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZOD
 
 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
6ZVJ
 
 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
2IX8
 
 | | MODEL FOR EEF3 BOUND TO AN 80S RIBOSOME | | Descriptor: | ELONGATION FACTOR 3A | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2J28
 
 | | MODEL OF E. COLI SRP BOUND TO 70S RNCS | | Descriptor: | 23S RIBOSOMAL RNA, 4.5S SIGNAL RECOGNITION PARTICLE RNA, 50S ribosomal protein L11, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Following the Signal Sequence from Ribosomal Tunnel Exit to Signal Recognition Particle
Nature, 444, 2006
|
|
2IW3
 
 | | Elongation Factor 3 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELONGATION FACTOR 3A, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2J37
 
 | | MODEL OF MAMMALIAN SRP BOUND TO 80S RNCS | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, RIBOSOMAL PROTEIN L31, RIBOSOMAL PROTEIN L35, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-18 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Following the signal sequence from ribosomal tunnel exit to signal recognition particle.
Nature, 444, 2006
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
2IX3
 
 | | Structure of yeast Elongation Factor 3 | | Descriptor: | ELONGATION FACTOR 3, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
6H4N
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
2IWH
 
 | | Structure of yeast Elongation Factor 3 in complex with ADPNP | | Descriptor: | ELONGATION FACTOR 3A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
6HD7
 
 | | Cryo-EM structure of the ribosome-NatA complex | | Descriptor: | 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Becker, T, Beckmann, R. | | Deposit date: | 2018-08-17 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
8I9T
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-1 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8I9P
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Mak16 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
