2WNX
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-20 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WO4
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum, in-house data | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XDH
 
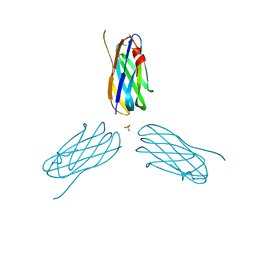 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
2VO8
 
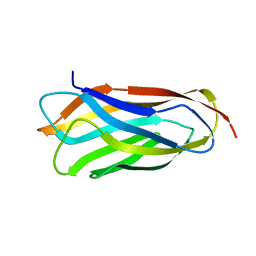 | | Cohesin module from Clostridium perfringens ATCC13124 family 33 glycoside hydrolase. | | Descriptor: | EXO-ALPHA-SIALIDASE | | Authors: | Gregg, K, Adams, J.J, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Clostridium Perfringens Toxin Complex Formation.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1LEL
 
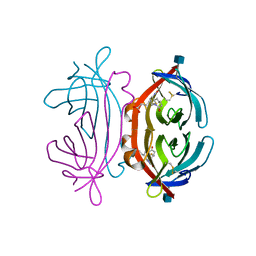 | | The avidin BCAP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, E-AMINO BIOTINYL CAPROIC ACID | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-10 | | Release date: | 2002-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1LCW
 
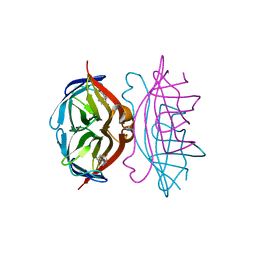 | | streptavidin-homobiotin complex | | Descriptor: | HOMOBIOTIN, Streptavidin | | Authors: | Livnah, O, Pazy, Y, Bayer, E.A, Wilchek, M. | | Deposit date: | 2002-04-07 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1LCZ
 
 | | streptavidin-BCAP complex | | Descriptor: | E-AMINO BIOTINYL CAPROIC ACID, Streptavidin | | Authors: | Livnah, O, Pazy, Y, Bayer, E.A, Wilchek, M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1LDO
 
 | | avidin-norbioitn complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NORBIOTIN, avidin | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-09 | | Release date: | 2002-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1LDQ
 
 | | avidin-homobiotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, HOMOBIOTIN | | Authors: | Pazy, Y, Kulik, T, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2002-04-09 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand exchange between proteins: exchange of biotin and biotin derivatives between avidin and streptavidin
J.Biol.Chem., 277, 2002
|
|
1RXH
 
 | | Crystal structure of streptavidin mutant L124R (M1) complexed with biotinyl p-nitroanilide (BNI) | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5OGZ
 
 | |
1RXK
 
 | | crystal structure of streptavidin mutant (M3) a combination of M1+M2 | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RXJ
 
 | | Crystal structure of streptavidin mutant (M2) where the L3,4 loop was replace by that of avidin | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2JH2
 
 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2XQO
 
 | | CtCel124: a cellulase from Clostridium thermocellum | | Descriptor: | Dockerin type 1, NICKEL (II) ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Carvalho, A.L, Verze, G, Bras, J.L.A, Cartmell, A, Bayer, E.A, Vazana, Y, Correia, M.A.S, Prates, J.A.M, Gilbert, H.J, Fontes, C.M.G.A, Romao, M.J. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights Into a Unique Cellulase Fold and Mechanism of Cellulose Hydrolysis
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XFG
 
 | | Reassembly and co-crystallization of a family 9 processive endoglucanase from separately expressed GH9 and CBM3c modules | | Descriptor: | CALCIUM ION, CHLORIDE ION, ENDOGLUCANASE 1 | | Authors: | Petkun, S, Lamed, R, Jindou, S, Burstein, T, Yaniv, O, Shoham, Y, Shimon, J.W.L, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Reassembly and Co-Crystallization of a Family 9 Processive Endoglucanase from its Component Parts: Structural and Functional Significance of Intermodular Linker
Peerj, 3, 2015
|
|
2YLK
 
 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1Y53
 
 | | Crystal structure of bacterial expressed avidin related protein 4 (AVR4) C122S | | Descriptor: | Avidin-related protein 4/5, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y52
 
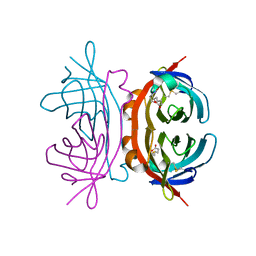 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y55
 
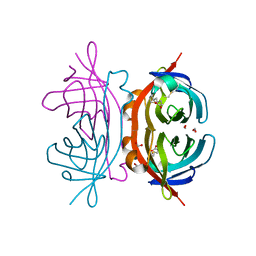 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | Descriptor: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
2JNK
 
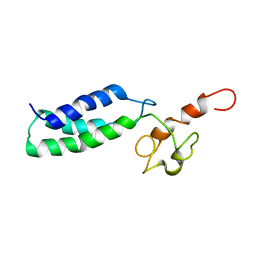 | |
2XBT
 
 | | Structure of a scaffoldin carbohydrate-binding module family 3b from the cellulosome of Bacteroides cellulosolvens: Structural diversity and implications for carbohydrate binding | | Descriptor: | CELLULOSOMAL SCAFFOLDIN, NITRATE ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2010-04-15 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Scaffoldin-Borne Family 3B Carbohydrate-Binding Module from the Cellulosome of Bacteroides Cellulosolvens: Structural Diversity and Significance of Calcium for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4B9P
 
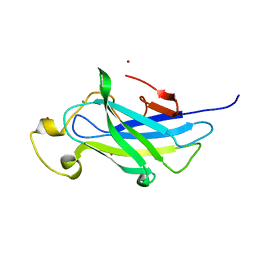 | | Biomass sensoring module from putative Rsgi2 protein of Clostridium thermocellum resemble family 3 carbohydrate-binding module of cellulosome | | Descriptor: | CALCIUM ION, TYPE 3A CELLULOSE-BINDING DOMAIN PROTEIN, ZINC ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Fine-Structural Variance of Family 3 Carbohydrate-Binding Modules as Extracellular Biomass-Sensing Components of Clostridium Thermocellum Anti-Sigma(I) Factors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4B9C
 
 | | Biomass sensoring modules from putative Rsgi-like proteins of Clostridium thermocellum resemble family 3 carbohydrate-binding module of cellulosome | | Descriptor: | CALCIUM ION, TYPE 3A CELLULOSE-BINDING DOMAIN PROTEIN | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Fine-Structural Variance of Family 3 Carbohydrate-Binding Modules as Extracellular Biomass-Sensing Components of Clostridium Thermocellum Anti-Sigma(I) Factors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
