1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
1NAJ
 
 | | High resolution NMR Structure Of DNA Dodecamer Determined In Aqueous Dilute Liquid Crystalline Phase | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Wu, Z, Delaglio, F, Tjandra, N, Zhurkin, V, Bax, A. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and (31)P chemical shift anisotropy.
J.Biomol.Nmr, 26, 2003
|
|
2JQX
 
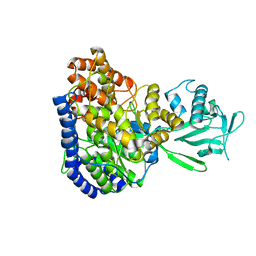 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | Descriptor: | Malate synthase G | | Authors: | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | Deposit date: | 2007-06-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | Descriptor: | 76-MER | | Authors: | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
2JWK
 
 | |
2JWL
 
 | |
2MIZ
 
 | |
2KXA
 
 | |
2LWA
 
 | | Conformational ensemble for the G8A mutant of the influenza hemagglutinin fusion peptide | | Descriptor: | HEMAGGLUTININ FUSION PEPTIDE G8A MUTANT | | Authors: | Lorieau, J.L, Louis, J.M, Schwieters, C.D, Bax, A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NEF
 
 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
2N7J
 
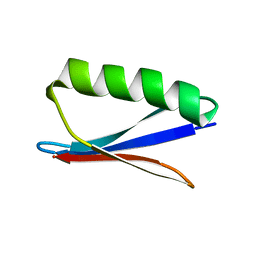 | |
2MJB
 
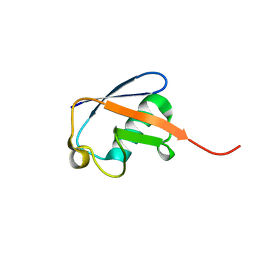 | | Solution nmr structure of ubiquitin refined against dipolar couplings in 4 media | | Descriptor: | Ubiquitin-60S ribosomal protein L40 | | Authors: | Maltsev, A, Grishaev, A, Roche, J, Zasloff, M, Bax, A. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved cross validation of a static ubiquitin structure derived from high precision residual dipolar couplings measured in a drug-based liquid crystalline phase.
J.Am.Chem.Soc., 136, 2014
|
|
2MK3
 
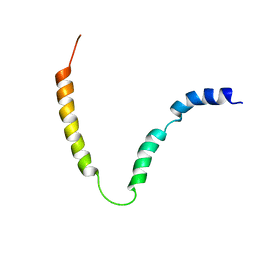 | | Solution NMR structure of gp41 ectodomain monomer on a DPC micelle | | Descriptor: | Transmembrane glycoprotein, chimeric construct | | Authors: | Roche, J, Louis, J.M, Grishaev, A, Ying, J, Bax, A. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OED
 
 | | GB3 solution structure obtained by refinement of X-ray structure with dipolar couplings | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ulmer, T.S, Ramirez, B.E, Delaglio, F, Bax, A, Grishaev, A. | | Deposit date: | 2006-12-29 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy
J.Am.Chem.Soc., 125, 2003
|
|
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | Descriptor: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | Authors: | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
4CQK
 
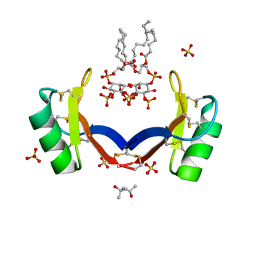 | | Crystal structure of ligand-bound NaD1 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLOWER-SPECIFIC DEFENSIN, SULFATE ION, ... | | Authors: | Lay, F.T, Mills, G.M, Poon, I.K.H, Baxter, A.A, Hulett, M.D, Kvansakul, M. | | Deposit date: | 2014-02-17 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphoinositide-Mediated Oligomerization of a Defensin Induces Cell Lysis.
Elife, 3, 2014
|
|
