5A0X
 
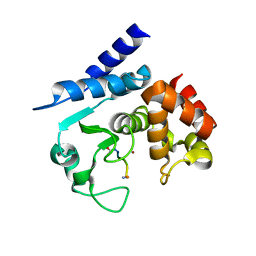 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | Descriptor: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0R
 
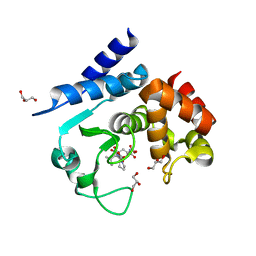 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
4Z8X
 
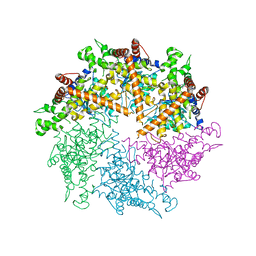 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, SULFATE ION, ... | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A0S
 
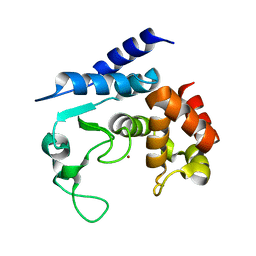 | | Apo-structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5OKT
 
 | | Crystal structure of human Casein Kinase I delta in complex with IWP-2 | | Descriptor: | ACETATE ION, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) delta / epsilon.
J. Med. Chem., 61, 2018
|
|
2VSO
 
 | | Crystal Structure of a Translation Initiation Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT RNA HELICASE EIF4A, EUKARYOTIC INITIATION FACTOR 4F SUBUNIT P150 | | Authors: | Schutz, P, Bumann, M, Oberholzer, A.E, Bieniossek, C, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Yeast Eif4A-Eif4G Complex: An RNA-Helicase Controlled by Protein-Protein Interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2XZ9
 
 | | CRYSTAL STRUCTURE FROM THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA), PYRUVIC ACID | | Authors: | Oberholzer, A.E, Waltersperger, S.M, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
2XZ7
 
 | | CRYSTAL STRUCTURE OF THE PHOSPHOENOLPYRUVATE-BINDING DOMAIN OF ENZYME I IN COMPLEX WITH PHOSPHOENOLPYRUVATE FROM THE THERMOANAEROBACTER TENGCONGENSIS PEP-SUGAR PHOSPHOTRANSFERASE SYSTEM (PTS) | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, PHOSPHOENOLPYRUVATE-PROTEIN KINASE (PTS SYSTEM EI COMPONENT IN BACTERIA) | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Schneider, P, Baumann, U, Erni, B. | | Deposit date: | 2010-11-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoenolpyruvate: Sugar Phosphotransferase System from the Hyperthermophilic Thermoanaerobacter Tengcongensis.
Biochemistry, 50, 2011
|
|
2WQF
 
 | | Crystal Structure of the Nitroreductase CinD from Lactococcus lactis in Complex with FMN | | Descriptor: | COPPER INDUCED NITROREDUCTASE D, FLAVIN MONONUCLEOTIDE | | Authors: | Waltersperger, S.M, Oberholzer, A.E, Solioz, M, Baumann, U. | | Deposit date: | 2009-08-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Cind (Ytjd) of Lactococcus Lactis, a Copper-Induced Nitroreductase Involved in Defense Against Oxidative Stress.
J.Bacteriol., 192, 2010
|
|
2WQD
 
 | | Crystal structure of enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system in the dephosphorylated state | | Descriptor: | CALCIUM ION, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Oberholzer, A.E, Schneider, P, Siebold, C, Baumann, U, Erni, B. | | Deposit date: | 2009-08-19 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Enzyme I of the Phosphoenolpyruvate:Sugar Phosphotransferase System in the Dephosphorylated State.
J.Biol.Chem., 284, 2009
|
|
2X7M
 
 | |
6HMP
 
 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azoimidazole-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6HMR
 
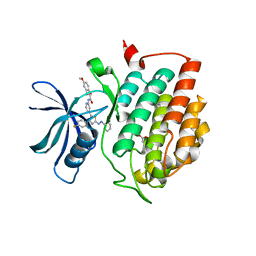 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azothiazole-based inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, MALONIC ACID | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5LNB
 
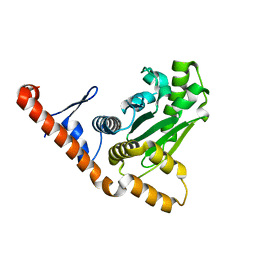 | |
5N12
 
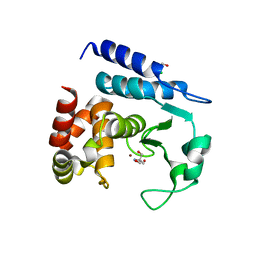 | | Crystal structure of TCE treated rPPEP-1 | | Descriptor: | 2,2,2-tris-chloroethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Schacherl, M, Baumann, U. | | Deposit date: | 2017-02-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1K7G
 
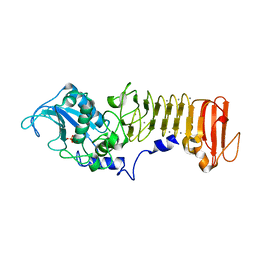 | | PrtC from Erwinia chrysanthemi | | Descriptor: | CALCIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Hege, T, Baumann, U. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1LO4
 
 | | Retro-Diels-Alderase Catalytic antibody 9D9 | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO0
 
 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | Descriptor: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-05 | | Release date: | 2002-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO2
 
 | | Retro-Diels-Alderase Catalytic Antibody | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain, [2'-CARBOXYLETHYL]-10-METHYL-ANTHRACENE ENDOPEROXIDE | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LO3
 
 | | Retro-Diels-Alderase Catalytic Antibody: Product Analogue | | Descriptor: | 3-(10-METHYL-ANTHRACEN-9-YL)-PROPIONIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1UOD
 
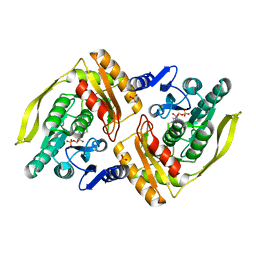 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with dihydroxyacetone-phosphate | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCERALDEHYDE-3-PHOSPHATE, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
1W2W
 
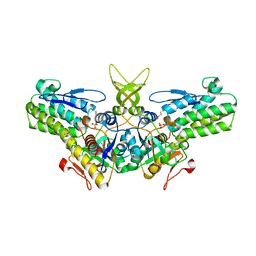 | | Crystal structure of yeast Ypr118w, a methylthioribose-1-phosphate isomerase related to regulatory eIF2B subunits | | Descriptor: | 5-METHYLTHIORIBOSE-1-PHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Bumann, M, Djafarzadeh, S, Oberholzer, A.E, Bigler, P, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Yeast Ypr118W, a Methylthioribose-1-Phosphate Isomerase Related to Regulatory Eif2B Subunits
J.Biol.Chem., 279, 2004
|
|
1UN8
 
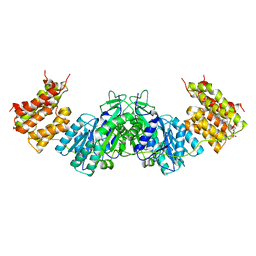 | | Crystal structure of the dihydroxyacetone kinase of C. freundii (native form) | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, DIHYDROXYACETONE KINASE | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
1UOE
 
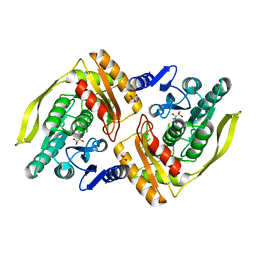 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with glyceraldehyde | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCEROL, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
1XFO
 
 | |
