2V3L
 
 | | Orientational and dynamical heterogeneity of Rhodamine 6G terminally attached to a DNA helix | | Descriptor: | 5'-D(*CP*AP*AP*AP*GP*CP*GP*CP*CP*AP *TP*TP*CP*GP*CP*CP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*GP*GP*CP*GP*AP*AP *TP*GP*GP*CP*GP*CP*TP*TP*TP*G)-3', RHODAMINE 6G | | Authors: | Neubauer, H, Gaiko, N, Berger, S, Schaffer, J, Eggeling, C, Tuma, J, Verdier, L, Seidel, C.A.M, Griesinger, C, Volkmer, A. | | Deposit date: | 2007-06-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Orientational and Dynamical Heterogeneity of Rhodamine 6G Terminally Attached to a DNA Helix Revealed by NMR and Single-Molecule Fluorescence Spectroscopy.
J.Am.Chem.Soc., 129, 2007
|
|
2GH5
 
 | | Crystal Structure of human Glutathione Reductase complexed with a Fluoro-Analogue of the Menadione Derivative M5 | | Descriptor: | 6-(3-METHYL-1,4-DIOXO-1,4-DIHYDRONAPHTHALEN-2-YL)HEXANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Winzer, A, Bauer, H, Schirmer, H, Davioud-Charvet, E. | | Deposit date: | 2006-03-25 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fluoro analogue of the menadione derivative 6-[2'-(3'-methyl)-1',4'-naphthoquinolyl]hexanoic acid is a suicide substrate of glutathione reductase. Crystal structure of the alkylated human enzyme
J.Am.Chem.Soc., 128, 2006
|
|
5FZP
 
 | | Structure of the dispase autolysis inducing protein from Streptomyces mobaraensis | | Descriptor: | CALCIUM ION, DISPASE AUTOLYSIS-INDUCING PROTEIN, GLYCEROL | | Authors: | Schmelz, S, Fiebig, D, Beck, J, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2016-03-15 | | Release date: | 2016-08-10 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
J.Biol.Chem., 291, 2016
|
|
6I0I
 
 | |
6TCX
 
 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
6FHP
 
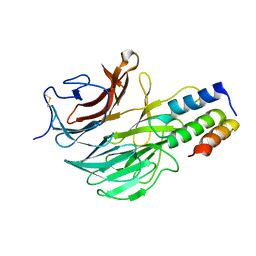 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
6GMG
 
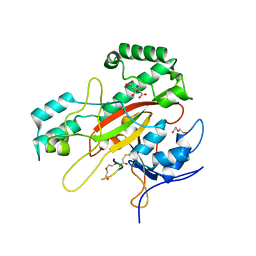 | | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, ... | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a glutamine donor mimicking inhibitory peptide shaped by the catalytic cleft of microbial transglutaminase.
FEBS J., 285, 2018
|
|
5NTB
 
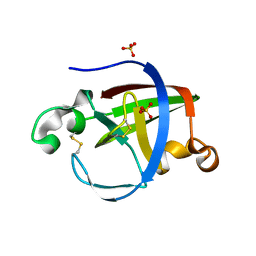 | | Streptomyces papain inhibitor (SPI) | | Descriptor: | Papain inhibitor, SULFATE ION | | Authors: | Schmelz, S, Juettner, N.E, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Illuminating structure and acyl donor sites of a physiological transglutaminase substrate from Streptomyces mobaraensis.
Protein Sci., 27, 2018
|
|
1NSO
 
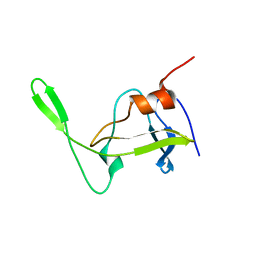 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
