395D
 
 | |
100D
 
 | |
161D
 
 | |
393D
 
 | |
394D
 
 | |
396D
 
 | |
279D
 
 | |
260D
 
 | |
2M8E
 
 | | NMR structure of the PAI subdomain of Sleeping Beauty transposase | | Descriptor: | SLEEPING BEAUTY TRANSPOSASE | | Authors: | Eubanks, C, Schreifels, J, Aronovich, E, Carlson, D, Hacjkett, P, Nesmelova, I. | | Deposit date: | 2013-05-17 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of Sleeping Beauty transposase binding to DNA.
Protein Sci., 23, 2014
|
|
315D
 
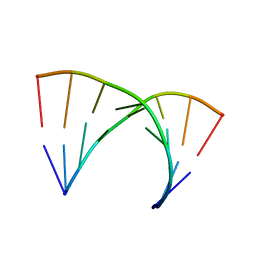 | |
1EWR
 
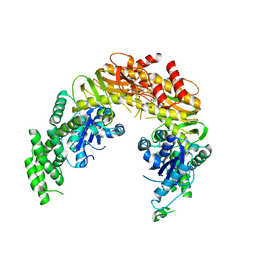 | | CRYSTAL STRUCTURE OF TAQ MUTS | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
1EWQ
 
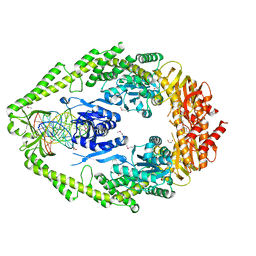 | | CRYSTAL STRUCTURE TAQ MUTS COMPLEXED WITH A HETERODUPLEX DNA AT 2.2 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3'), ... | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
4HN7
 
 | |
3HR8
 
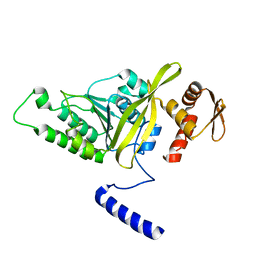 | | Crystal Structure of Thermotoga maritima RecA | | Descriptor: | Protein recA | | Authors: | Lee, S, Kim, T.G, Jeong, E.-Y, Ban, C, Jeon, W.-J, Min, K.I, Song, K.-M, Heo, S.-D, Ku, J.K. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of RecA Protein from Thermotoga maritima MSB8
to be published
|
|
2J7Z
 
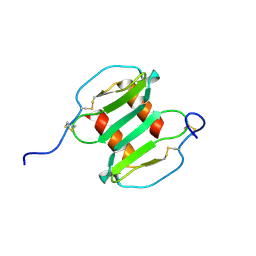 | | Crystal Structure of recombinant Human Stromal Cell-Derived Factor- 1alpha | | Descriptor: | STROMAL CELL-DERIVED FACTOR 1 ALPHA | | Authors: | Ryu, E.K, Kim, T.G, Kwon, T.H, Jung, I.D, Ryu, D.W, Park, Y.-M, Ahn, K, Ban, C. | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Recombinant Human Stromal Cell-Derived Factor-1Alpha.
Proteins, 67, 2007
|
|
5HTO
 
 | |
5HS4
 
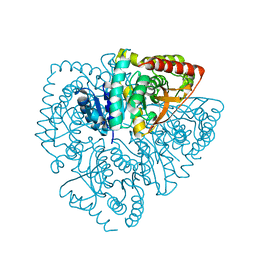 | | Plasmdoium Vivax Lactate dehydrogenase | | Descriptor: | L-lactate dehydrogenase | | Authors: | Choi, S.J, Ban, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition
Sci Rep, 6, 2016
|
|
5HRU
 
 | |
246D
 
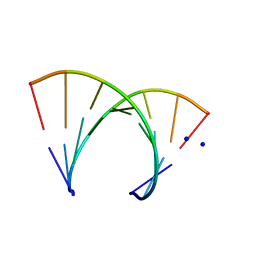 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1BKN
 
 | |
331D
 
 | | CRYSTAL STRUCTURE OF D(GCGCGCG) WITH 5'-OVERHANG G'S | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*GP*CP*GP*CP*G)-3') | | Authors: | Pan, B, Ban, C, Wahl, M, Sundaralingam, M. | | Deposit date: | 1997-05-13 | | Release date: | 1997-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of d(GCGCGCG) with 5'-overhang G residues.
Biophys.J., 73, 1997
|
|
1KNV
 
 | | Bse634I restriction endonuclease | | Descriptor: | ACETATE ION, Bse634I restriction endonuclease, CHLORIDE ION | | Authors: | Grazulis, S, Deibert, M, Rimseliene, R, Skirgaila, R, Sasnauskas, G, Lagunavicius, A, Repin, V, Urbanke, C, Huber, R, Siksnys, V. | | Deposit date: | 2001-12-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence.
Nucleic Acids Res., 30, 2002
|
|
2N9H
 
 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9F
 
 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3ULL
 
 | |
