6ND1
 
 | | CryoEM structure of the Sec Complex from yeast | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Wu, X, Cabanos, C, Rapoport, T.A. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the post-translational protein translocation machinery of the ER membrane.
Nature, 566, 2019
|
|
2BPM
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-630529 | | Descriptor: | (2S)-N-[(3Z)-5-CYCLOPROPYL-3H-PYRAZOL-3-YLIDENE]-2-[4-(2-OXOIMIDAZOLIDIN-1-YL)PHENYL]PROPANAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cameron, A, Fogliatto, G, Pevarello, P, Brasca, M.G, Orsini, P, Traquandi, G, Longo, A, Nesi, M, Orzi, F, Piutti, C, Sansonna, P, Varasi, M, Vulpetti, A, Roletto, F, Alzani, R, Ciomei, M, Albanese, C, Pastori, W, Marsiglio, A, Pesenti, E, Fiorentini, F, Bischoff, J.R, Mercurio, C. | | Deposit date: | 2005-04-21 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2-Cyclin a as Antitumor Agents. 2. Lead Optimization
J.Med.Chem., 48, 2005
|
|
3KSC
 
 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | Descriptor: | GLYCEROL, LegA class, SULFATE ION | | Authors: | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-11-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
6BIH
 
 | | The Structure of the Actin-Smooth Muscle Myosin Motor Domain Complex in the Rigor State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Taylor, K.A, Banerjee, C, Hu, Z. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The structure of the actin-smooth muscle myosin motor domain complex in the rigor state.
J. Struct. Biol., 200, 2017
|
|
2WPA
 
 | | Optimisation of 6,6-Dimethyl Pyrrolo 3,4-c pyrazoles: Identification of PHA-793887, a Potent CDK Inhibitor Suitable for Intravenous Dosing | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-{6,6-DIMETHYL-5-[(1-METHYLPIPERIDIN-4-YL)CARBONYL]-1,4,5,6-TETRAHYDROPYRROLO[3,4-C]PYRAZOL-3-YL}-3-METHYLBUTANAMIDE, ... | | Authors: | Brasca, M.G, Albanese, C, Alzani, R, Amici, R, Avanzi, N, Ballinari, D, Bischoff, J, Borghi, D, Casale, E, Croci, V, Fiorentini, F, Isacchi, A, Mercurio, C, Nesi, M, Orsini, P, Pastori, W, Pesenti, E, Pevarello, P, Roussel, P, Varasi, M, Volpi, D, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Optimization of 6,6-Dimethyl Pyrrolo[3,4-C]Pyrazoles: Identification of Pha-793887, a Potent Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem., 18, 2010
|
|
4A0F
 
 | | Structure of selenomethionine substituted bifunctional DAPA aminotransferase-dethiobiotin synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.714 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0G
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0H
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to 7-keto 8-amino pelargonic acid (KAPA) | | Descriptor: | 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0R
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to dethiobiotin (DTB). | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
1WA9
 
 | | Crystal Structure of the PAS repeat region of the Drosophila clock protein PERIOD | | Descriptor: | PERIOD CIRCADIAN PROTEIN | | Authors: | Yildiz, O, Doi, M, Yujnovsky, I, Cardone, L, Berndt, A, Hennig, S, Schulze, S, Urbanke, C, Sassone-Corsi, P, Wolf, E. | | Deposit date: | 2004-10-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Mol.Cell, 17, 2005
|
|
1O6T
 
 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-15 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
2Q10
 
 | | RESTRICTION ENDONUCLEASE BcnI (WILD TYPE)-COGNATE DNA SUBSTRATE COMPLEX | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3'), ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2007-05-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Monomeric Restriction Endonuclease BcnI in the Apo Form and in an Asymmetric Complex with Target DNA.
J.Mol.Biol., 369, 2007
|
|
1O6S
 
 | | Internalin (Listeria monocytogenes) / E-Cadherin (human) Recognition Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, E-CADHERIN, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-13 | | Release date: | 2002-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1O6V
 
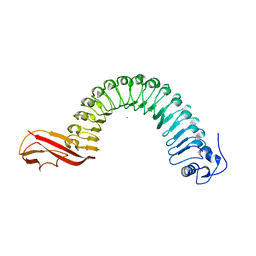 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | CALCIUM ION, INTERNALIN A | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
2IHE
 
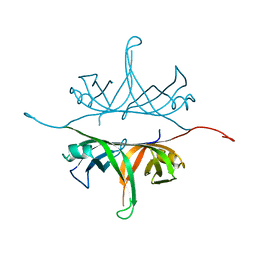 | | Crystal structure of wild-type single-stranded DNA binding protein from Thermus aquaticus | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Fedorov, R, Witte, G, Urbanke, C, Manstein, D.J, Curth, U. | | Deposit date: | 2006-09-26 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins.
Nucleic Acids Res., 34, 2006
|
|
2IXS
 
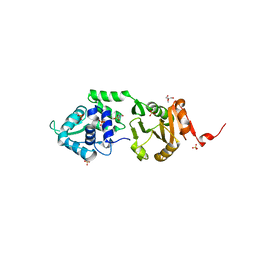 | | Structure of SdaI restriction endonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SDAI RESTRICTION ENDONUCLEASE, ... | | Authors: | Tamulaitiene, G, Jakubauskas, A, Urbanke, C, Huber, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Rare-Cutting Restriction Enzyme Sdai Reveals Unexpected Domain Architecture
Structure, 14, 2006
|
|
5ZGG
 
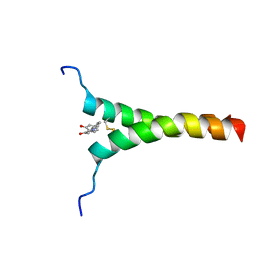 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
2IHF
 
 | | Crystal structure of deletion mutant delta 228-252 R190A of the single-stranded DNA binding protein from Thermus aquaticus | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Fedorov, R, Witte, G, Urbanke, C, Manstein, D.J, Curth, U. | | Deposit date: | 2006-09-26 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins.
Nucleic Acids Res., 34, 2006
|
|
2ODI
 
 | | Restriction Endonuclease BCNI-Cognate DNA Substrate Complex | | Descriptor: | 5'-D(*AP*AP*CP*CP*CP*GP*GP*AP*GP*AP*C)-3', 5'-D(*CP*TP*CP*CP*GP*GP*GP*TP*TP*GP*T)-3', CALCIUM ION, ... | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, C, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
